Recurrent Loss of APOBEC3H Activity during Primate Evolution
- PMID: 29925657
- PMCID: PMC6096791
- DOI: 10.1128/JVI.00971-18
Recurrent Loss of APOBEC3H Activity during Primate Evolution
Abstract
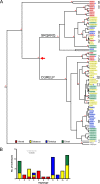
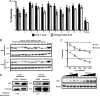
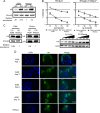
Genes in the APOBEC3 family encode cytidine deaminases that provide a barrier against viral infection and retrotransposition. Of all the APOBEC3 genes in humans, APOBEC3H (A3H) is the most polymorphic: some genes encode stable and active A3H proteins, while others are unstable and poorly antiviral. Such variation in human A3H affects interactions with the lentiviral antagonist Vif, which counteracts A3H via proteasomal degradation. In order to broaden our understanding of A3H-Vif interactions, as well as its evolution in Old World monkeys, we characterized A3H variation within four African green monkey (AGM) subspecies. We found that A3H is highly polymorphic in AGMs and has lost antiviral activity in multiple Old World monkeys. This loss of function was partially related to protein expression levels but was also influenced by amino acid mutations in the N terminus. Moreover, we demonstrate that the evolution of A3H in the primate lineages leading to AGMs was not driven by Vif. Our work suggests that the activity of A3H is evolutionarily dynamic and may have a negative effect on host fitness, resulting in its recurrent loss in primates.IMPORTANCE Adaptation of viruses to their hosts is critical for viral transmission between different species. Previous studies had identified changes in a protein from the APOBEC3 family that influenced the species specificity of simian immunodeficiency viruses (SIVs) in African green monkeys. We studied the evolution of a related protein in the same system, APOBEC3H, which has experienced a loss of function in humans. This evolutionary approach revealed that recurrent loss of APOBEC3H activity has taken place during primate evolution, suggesting that APOBEC3H places a fitness cost on hosts. The variability of APOBEC3H activity between different primates highlights the differential selective pressures on the APOBEC3 gene family.
Keywords: APOBEC3H; African green monkey; evolution; human immunodeficiency virus; innate immunity; lentiviruses.
Copyright © 2018 American Society for Microbiology.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

