Bacterial RecA Protein Promotes Adenoviral Recombination during In Vitro Infection
- PMID: 29925671
- PMCID: PMC6010623
- DOI: 10.1128/mSphere.00105-18
Bacterial RecA Protein Promotes Adenoviral Recombination during In Vitro Infection
Abstract
Adenovirus infections in humans are common and sometimes lethal. Adenovirus-derived vectors are also commonly chosen for gene therapy in human clinical trials. We have shown in previous work that homologous recombination between adenoviral genomes of human adenovirus species D (HAdV-D), the largest and fastest growing HAdV species, is responsible for the rapid evolution of this species. Because adenovirus infection initiates in mucosal epithelia, particularly at the gastrointestinal, respiratory, genitourinary, and ocular surfaces, we sought to determine a possible role for mucosal microbiota in adenovirus genome diversity. By analysis of known recombination hot spots across 38 human adenovirus genomes in species D (HAdV-D), we identified nucleotide sequence motifs similar to bacterial Chi sequences, which facilitate homologous recombination in the presence of bacterial Rec enzymes. These motifs, referred to here as ChiAD, were identified immediately 5' to the sequence encoding penton base hypervariable loop 2, which expresses the arginine-glycine-aspartate moiety critical to adenoviral cellular entry. Coinfection with two HAdV-Ds in the presence of an Escherichia coli lysate increased recombination; this was blocked in a RecA mutant strain, E. coli DH5α, or upon RecA depletion. Recombination increased in the presence of E. coli lysate despite a general reduction in viral replication. RecA colocalized with viral DNA in HAdV-D-infected cell nuclei and was shown to bind specifically to ChiAD sequences. These results indicate that adenoviruses may repurpose bacterial recombination machinery, a sharing of evolutionary mechanisms across a diverse microbiota, and unique example of viral commensalism.IMPORTANCE Adenoviruses are common human mucosal pathogens of the gastrointestinal, respiratory, and genitourinary tracts and ocular surface. Here, we report finding Chi-like sequences in adenovirus recombination hot spots. Adenovirus coinfection in the presence of bacterial RecA protein facilitated homologous recombination between viruses. Genetic recombination led to evolution of an important external feature on the adenoviral capsid, namely, the penton base protein hypervariable loop 2, which contains the arginine-glycine-aspartic acid motif critical to viral internalization. We speculate that free Rec proteins present in gastrointestinal secretions upon bacterial cell death facilitate the evolution of human adenoviruses through homologous recombination, an example of viral commensalism and the complexity of virus-host interactions, including regional microbiota.
Keywords: adenoviruses; commensal; homologous recombination.
Copyright © 2018 Lee et al.
Figures

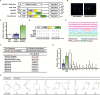
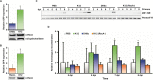
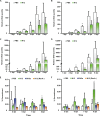
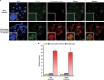
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
