Shift from stochastic to spatially-ordered expression of serine-glycine synthesis enzymes in 3D microtumors
- PMID: 29925909
- PMCID: PMC6010463
- DOI: 10.1038/s41598-018-27266-8
Shift from stochastic to spatially-ordered expression of serine-glycine synthesis enzymes in 3D microtumors
Abstract
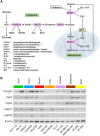
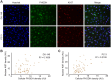
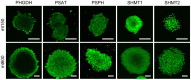
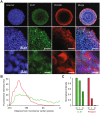
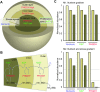
Cell-to-cell differences in protein expression in normal tissues and tumors are a common phenomenon, but the underlying principles that govern this heterogeneity are largely unknown. Here, we show that in monolayer cancer cell-line cultures, the expression of the five metabolic enzymes of serine-glycine synthesis (SGS), including its rate-limiting enzyme, phosphoglycerate dehydrogenase (PHGDH), displays stochastic cell-to-cell variation. By contrast, in cancer cell line-derived three-dimensional (3D) microtumors PHGDH expression is restricted to the outermost part of the microtumors' outer proliferative cell layer, while the four other SGS enzymes display near uniform expression throughout the microtumor. A mathematical model suggests that metabolic stress in the microtumor core activates factors that restrict PHGDH expression. Thus, intracellular enzyme expression in growing cell ecosystems can shift to spatially ordered patterns in 3D structured environments due to emergent cell-cell communication, with potential implications for the design of effective anti-metabolic cancer therapies.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

