A multiprotein supercomplex controlling oncogenic signalling in lymphoma
- PMID: 29925955
- PMCID: PMC6201842
- DOI: 10.1038/s41586-018-0290-0
A multiprotein supercomplex controlling oncogenic signalling in lymphoma
Abstract
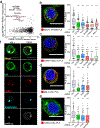
B cell receptor (BCR) signalling has emerged as a therapeutic target in B cell lymphomas, but inhibiting this pathway in diffuse large B cell lymphoma (DLBCL) has benefited only a subset of patients1. Gene expression profiling identified two major subtypes of DLBCL, known as germinal centre B cell-like and activated B cell-like (ABC)2,3, that show poor outcomes after immunochemotherapy in ABC. Autoantigens drive BCR-dependent activation of NF-κB in ABC DLBCL through a kinase signalling cascade of SYK, BTK and PKCβ to promote the assembly of the CARD11-BCL10-MALT1 adaptor complex, which recruits and activates IκB kinase4-6. Genome sequencing revealed gain-of-function mutations that target the CD79A and CD79B BCR subunits and the Toll-like receptor signalling adaptor MYD885,7, with MYD88(L265P) being the most prevalent isoform. In a clinical trial, the BTK inhibitor ibrutinib produced responses in 37% of cases of ABC1. The most striking response rate (80%) was observed in tumours with both CD79B and MYD88(L265P) mutations, but how these mutations cooperate to promote dependence on BCR signalling remains unclear. Here we used genome-wide CRISPR-Cas9 screening and functional proteomics to determine the molecular basis of exceptional clinical responses to ibrutinib. We discovered a new mode of oncogenic BCR signalling in ibrutinib-responsive cell lines and biopsies, coordinated by a multiprotein supercomplex formed by MYD88, TLR9 and the BCR (hereafter termed the My-T-BCR supercomplex). The My-T-BCR supercomplex co-localizes with mTOR on endolysosomes, where it drives pro-survival NF-κB and mTOR signalling. Inhibitors of BCR and mTOR signalling cooperatively decreased the formation and function of the My-T-BCR supercomplex, providing mechanistic insight into their synergistic toxicity for My-T-BCR+ DLBCL cells. My-T-BCR supercomplexes characterized ibrutinib-responsive malignancies and distinguished ibrutinib responders from non-responders. Our data provide a framework for the rational design of oncogenic signalling inhibitors in molecularly defined subsets of DLBCL.
Conflict of interest statement
The authors state they have no competing interests.
Figures








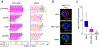




References
-
- Alizadeh AA et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511 (2000). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

