Time-Resolved Hydroxyl Radical Footprinting of RNA with X-Rays
- PMID: 29927103
- PMCID: PMC6057793
- DOI: 10.1002/cpnc.52
Time-Resolved Hydroxyl Radical Footprinting of RNA with X-Rays
Abstract
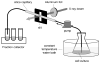
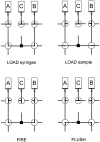
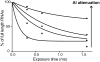
RNA footprinting by hydroxyl radical cleavage provides 'snapshots' of RNA tertiary structure or protein interactions that bury the RNA backbone. Generation of hydroxyl radicals with a high-flux synchrotron X-ray beam provides analysis on a short timescale (5-100 msec), which enables the structures of folding intermediates or other transient conformational states to be determined in biochemical solutions or cells. This article provides protocols for using synchrotron beamlines for hydroxyl radical footprinting. © 2018 by John Wiley & Sons, Inc.
Keywords: RNA structure probing; hydroxyl radical footprinting; time-resolved footprinting.
© 2018 John Wiley & Sons, Inc.
Figures


Similar articles
-
Time-resolved hydroxyl radical footprinting of RNA with X-rays.Curr Protoc Nucleic Acid Chem. 2001 Nov;Chapter 11:Unit 11.6. doi: 10.1002/0471142700.nc1106s06. Curr Protoc Nucleic Acid Chem. 2001. PMID: 18428832
-
Structural analysis of RNA in living cells by in vivo synchrotron X-ray footprinting.Methods Enzymol. 2009;468:239-58. doi: 10.1016/S0076-6879(09)68012-5. Methods Enzymol. 2009. PMID: 20946773 Free PMC article.
-
Time-resolved synchrotron X-ray "footprinting", a new approach to the study of nucleic acid structure and function: application to protein-DNA interactions and RNA folding.J Mol Biol. 1997 Feb 14;266(1):144-59. doi: 10.1006/jmbi.1996.0775. J Mol Biol. 1997. PMID: 9054977
-
Recent Advances and Applications in Synchrotron X-Ray Protein Footprinting for Protein Structure and Dynamics Elucidation.Protein Pept Lett. 2016;23(3):309-22. doi: 10.2174/0929866523666160201150057. Protein Pept Lett. 2016. PMID: 26833224 Review.
-
Probing the structural dynamics of nucleic acids by quantitative time-resolved and equilibrium hydroxyl radical "footprinting".Curr Opin Struct Biol. 2002 Oct;12(5):648-53. doi: 10.1016/s0959-440x(02)00366-4. Curr Opin Struct Biol. 2002. PMID: 12464318 Review.
Cited by
-
Role of Era in assembly and homeostasis of the ribosomal small subunit.Nucleic Acids Res. 2019 Sep 5;47(15):8301-8317. doi: 10.1093/nar/gkz571. Nucleic Acids Res. 2019. PMID: 31265110 Free PMC article.
-
Advances that facilitate the study of large RNA structure and dynamics by nuclear magnetic resonance spectroscopy.Wiley Interdiscip Rev RNA. 2019 Sep;10(5):e1541. doi: 10.1002/wrna.1541. Epub 2019 Apr 25. Wiley Interdiscip Rev RNA. 2019. PMID: 31025514 Free PMC article. Review.
-
New high-throughput endstation to accelerate the experimental optimization pipeline for synchrotron X-ray footprinting.J Synchrotron Radiat. 2021 Sep 1;28(Pt 5):1321-1332. doi: 10.1107/S1600577521005026. Epub 2021 Jul 20. J Synchrotron Radiat. 2021. PMID: 34475281 Free PMC article.
-
Phase separation and viral factories: unveiling the physical processes supporting RNA packaging in dsRNA viruses.Biochem Soc Trans. 2024 Oct 30;52(5):2101-2112. doi: 10.1042/BST20231304. Biochem Soc Trans. 2024. PMID: 39324618 Free PMC article. Review.
-
Evolution of a virus-like architecture and packaging mechanism in a repurposed bacterial protein.Science. 2021 Jun 11;372(6547):1220-1224. doi: 10.1126/science.abg2822. Science. 2021. PMID: 34112695 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

