Quantification of bacterial fluorescence using independent calibrants
- PMID: 29928012
- PMCID: PMC6013168
- DOI: 10.1371/journal.pone.0199432
Quantification of bacterial fluorescence using independent calibrants
Abstract
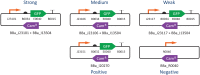
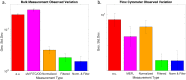
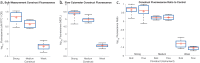
Fluorescent reporters are commonly used to quantify activities or properties of both natural and engineered cells. Fluorescence is still typically reported only in arbitrary or normalized units, however, rather than in units defined using an independent calibrant, which is problematic for scientific reproducibility and even more so when it comes to effective engineering. In this paper, we report an interlaboratory study showing that simple, low-cost unit calibration protocols can remedy this situation, producing comparable units and dramatic improvements in precision over both arbitrary and normalized units. Participants at 92 institutions around the world measured fluorescence from E. coli transformed with three engineered test plasmids, plus positive and negative controls, using simple, low-cost unit calibration protocols designed for use with a plate reader and/or flow cytometer. In addition to providing comparable units, use of an independent calibrant allows quantitative use of positive and negative controls to identify likely instances of protocol failure. The use of independent calibrants thus allows order of magnitude improvements in precision, narrowing the 95% confidence interval of measurements in our study up to 600-fold compared to normalized units.
Conflict of interest statement
The authors of this manuscript have read the journal’s policy and have the following competing interests: The authors received no specific commercial funding for this work. The following authors are employed by for-profit companies: Jacob Beal is employed by Raytheon BBN Technologies; Markus Gershater is employed by Synthace, and their work on this paper was thus indirectly supported by their salaries. This does not alter the authors’ adherence to PLOS ONE policies on sharing data and materials.
Figures

References
-
- Vogt RF Jr, Marti GE, Zenger V. Quantitative fluorescence calibration: a tool for assessing the quality of data obtained by fluorescence measurements In: Standardization and Quality Assurance in Fluorescence Measurements I. Springer; 2008. p. 3–31.
-
- Hoffman RA, Wang L, Bigos M, Nolan JP. NIST/ISAC standardization study: Variability in assignment of intensity values to fluorescence standard beads and in cross calibration of standard beads to hard dyed beads. Cytometry Part A. 2012;81(9):785–796. doi: 10.1002/cyto.a.22086 - DOI - PubMed
-
- Kelly JR, Rubin AJ, Davis JH, Ajo-Franklin CM, Cumbers J, Czar MJ, et al. Measuring the activity of BioBrick promoters using an in vivo reference standard. Journal of Biological Engineering. 2009;3(4). doi: 10.1186/1754-1611-3-4 - DOI - PMC - PubMed
-
- Yordanov B, Dalchau N, Grant PK, Pedersen M, Emmott S, Haseloff J, et al. A computational method for automated characterization of genetic components. ACS synthetic biology. 2014;3(8):578–588. doi: 10.1021/sb400152n - DOI - PubMed
-
- Stanton BC, Nielsen AA, Tamsir A, Clancy K, Peterson T, Voigt C. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nature Chemical Biology. 2014;10(2):99–105. doi: 10.1038/nchembio.1411 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

