Egr-1: A Candidate Transcription Factor Involved in Molecular Processes Underlying Time-Memory
- PMID: 29928241
- PMCID: PMC5997935
- DOI: 10.3389/fpsyg.2018.00865
Egr-1: A Candidate Transcription Factor Involved in Molecular Processes Underlying Time-Memory
Abstract
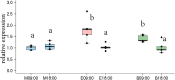
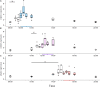
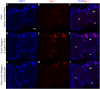
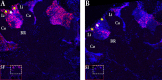
In honey bees, continuous foraging is accompanied by a sustained up-regulation of the immediate early gene Egr-1 (early growth response protein-1) and candidate downstream genes involved in learning and memory. Here, we present a series of feeder training experiments indicating that Egr-1 expression is highly correlated with the time and duration of training even in the absence of the food reward. Foragers that were trained to visit a feeder over the whole day and then collected on a day without food presentation showed Egr-1 up-regulation over the whole day with a peak expression around 14:00. When exposed to a time-restricted feeder presentation, either 2 h in the morning or 2 h in the evening, Egr-1 expression in the brain was up-regulated only during the hours of training. Foragers that visited a feeder in the morning as well as in the evening showed two peaks of Egr-1 expression. Finally, when we prevented time-trained foragers from leaving the colony using artificial rain, Egr-1 expression in the brains was still slightly but significantly up-regulated around the time of feeder training. In situ hybridization studies showed that active foraging and time-training induced Egr-1 up-regulation occurred in the same brain areas, preferentially the small Kenyon cells of the mushroom bodies and the antennal and optic lobes. Based on these findings we propose that foraging induced Egr-1 expression can get regulated by the circadian clock after time-training over several days and Egr-1 is a candidate transcription factor involved in molecular processes underlying time-memory.
Keywords: Egr-1; anticipation; honey bee foraging; small Kenyon cells; time-memory.
Figures


Similar articles
-
Honey bee foraging induces upregulation of early growth response protein 1, hormone receptor 38 and candidate downstream genes of the ecdysteroid signalling pathway.Insect Mol Biol. 2018 Feb;27(1):90-98. doi: 10.1111/imb.12350. Epub 2017 Oct 7. Insect Mol Biol. 2018. PMID: 28987007
-
High experience levels delay recruitment but promote simultaneous time-memories in honey bee foragers.J Exp Biol. 2018 Nov 26;221(Pt 23):jeb187336. doi: 10.1242/jeb.187336. J Exp Biol. 2018. PMID: 30337357
-
Neurogenomic signatures of spatiotemporal memories in time-trained forager honey bees.J Exp Biol. 2011 Mar 15;214(Pt 6):979-87. doi: 10.1242/jeb.053421. J Exp Biol. 2011. PMID: 21346126 Free PMC article.
-
Genetics in the Honey Bee: Achievements and Prospects toward the Functional Analysis of Molecular and Neural Mechanisms Underlying Social Behaviors.Insects. 2019 Oct 16;10(10):348. doi: 10.3390/insects10100348. Insects. 2019. PMID: 31623209 Free PMC article. Review.
-
Juvenile hormone, behavioral maturation, and brain structure in the honey bee.Dev Neurosci. 1996;18(1-2):102-14. doi: 10.1159/000111474. Dev Neurosci. 1996. PMID: 8840089 Review.
Cited by
-
Immediate early genes in social insects: a tool to identify brain regions involved in complex behaviors and molecular processes underlying neuroplasticity.Cell Mol Life Sci. 2019 Feb;76(4):637-651. doi: 10.1007/s00018-018-2948-z. Epub 2018 Oct 22. Cell Mol Life Sci. 2019. PMID: 30349993 Free PMC article. Review.
-
Search Behavior of Individual Foragers Involves Neurotransmitter Systems Characteristic for Social Scouting.Front Insect Sci. 2021 Jun 4;1:664978. doi: 10.3389/finsc.2021.664978. eCollection 2021. Front Insect Sci. 2021. PMID: 38468879 Free PMC article.
-
In search of behavioral and brain processes involved in honey bee dance communication.Front Behav Neurosci. 2023 Jun 29;17:1140657. doi: 10.3389/fnbeh.2023.1140657. eCollection 2023. Front Behav Neurosci. 2023. PMID: 37456809 Free PMC article. Review.
-
The Neural Signature of Visual Learning Under Restrictive Virtual-Reality Conditions.Front Behav Neurosci. 2022 Feb 16;16:846076. doi: 10.3389/fnbeh.2022.846076. eCollection 2022. Front Behav Neurosci. 2022. PMID: 35250505 Free PMC article.
-
Sociality sculpts similar patterns of molecular evolution in two independently evolved lineages of eusocial bees.Commun Biol. 2021 Feb 26;4(1):253. doi: 10.1038/s42003-021-01770-6. Commun Biol. 2021. PMID: 33637860 Free PMC article.
References
-
- Beier W. (1968). Beeinflussung der inneren Uhr der Bienen durch Phasenverschiebung des Licht–Dunkel–Zeitgebers. Zeitschrift Bienenforschung 9 356–378.
-
- Beling I. (1929). Über das Zeitgedächtnis der Bienen. Z. Vergl. Physiol. 9 259–338. 10.1007/BF00340159 - DOI
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B 57 289–300. 10.2307/2346101 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

