Genetic Diversity of Bovine Major Histocompatibility Complex Class II DRB3 locus in cattle breeds from Asia compared to those from Africa and America
- PMID: 29928467
- PMCID: PMC6004549
- DOI: 10.7150/jgen.26491
Genetic Diversity of Bovine Major Histocompatibility Complex Class II DRB3 locus in cattle breeds from Asia compared to those from Africa and America
Abstract
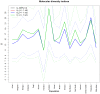
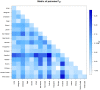
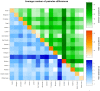
Genetic polymorphisms and diversity of BoLA-DRB3.2 are essential because of DRB3 gene's function in innate immunity and its association with infectious diseases resistance or tolerance in cattle. The present study was aimed at assessing the level of genetic diversity of DRB3 in the exon 2 (BoLA-DRB3.2) region in African, American and Asian cattle breeds. Amplification of exon 2 in 174 cattle revealed 15 haplotypes. The breeds with the highest number of haplotypes were Brangus (10), Sokoto Gudali (10) and Dajal (9), while the lowest number of haplotypes were found in Holstein and Sahiwal with 4 haplotypes each. Medium Joining network obtained from haplotypic data showed that all haplotypes condensed around a centric area and each sequence (except in H-3, H-51 and H-106) representing almost a specific haplotype. The BoLA-DRB3.2 sequence analyses revealed a non-significant higher rate of non-synonymous (dN) compared to synonymous substitutions (dS). The ratio of dN/dS substitution across the breeds were observed to be greater than one suggesting that variation at the antigen-binding sites is under positive selection; thus increasing the chances of these breeds to respond to wide array of pathogenic attacks. An analysis of molecular variance revealed that 94.01 and 5.99% of the genetic variation was attributable to differences within and among populations, respectively. Generally, results obtained suggest that within breed genetic variation across breeds is higher than between breeds. This genetic information will be important for investigating the relationship between BoLADRB3.2 and diseases in various cattle breeds studied with attendant implication on designing breeding programs that will aim at selecting individual cattle that carry resistant alleles.
Keywords: BoLA-DRB3.2; Cattle; Diseases; Gene polymorphism.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



Similar articles
-
Characterization of bovine MHC DRB3 diversity in global cattle breeds, with a focus on cattle in Myanmar.BMC Genet. 2020 Sep 1;21(1):95. doi: 10.1186/s12863-020-00905-8. BMC Genet. 2020. PMID: 32867670 Free PMC article.
-
The great diversity of major histocompatibility complex class II genes in Philippine native cattle.Meta Gene. 2014 Feb 19;2:176-90. doi: 10.1016/j.mgene.2013.12.005. eCollection 2014 Dec. Meta Gene. 2014. PMID: 25606401 Free PMC article.
-
Genetic diversity of BoLA-DRB3 in South American Zebu cattle populations.BMC Genet. 2018 May 22;19(1):33. doi: 10.1186/s12863-018-0618-7. BMC Genet. 2018. PMID: 29788904 Free PMC article.
-
Graduate Student Literature Review: The DRB3 gene of the bovine major histocompatibility complex-Discovery, diversity, and distribution of alleles in commercial breeds of cattle and applications for development of vaccines.J Dairy Sci. 2024 Dec;107(12):11324-11341. doi: 10.3168/jds.2023-24628. Epub 2024 Jul 14. J Dairy Sci. 2024. PMID: 39004123 Review.
-
Genetic variation and responses to vaccines.Anim Health Res Rev. 2004 Dec;5(2):197-208. doi: 10.1079/ahr200469. Anim Health Res Rev. 2004. PMID: 15984325 Review.
Cited by
-
Evolutionary Pattern of Interferon Alpha Genes in Bovidae and Genetic Diversity of IFNAA in the Bovine Genome.Front Immunol. 2020 Sep 30;11:580412. doi: 10.3389/fimmu.2020.580412. eCollection 2020. Front Immunol. 2020. PMID: 33117386 Free PMC article.
-
Genetic assessment of BoLA-DRB3 polymorphisms by comparing Bangladesh, Ethiopian, and Korean cattle.J Anim Sci Technol. 2021 Mar;63(2):248-261. doi: 10.5187/jast.2021.e37. Epub 2021 Mar 31. J Anim Sci Technol. 2021. PMID: 33987601 Free PMC article.
References
-
- Alvarez-Busto J, García-Etxebarria K, Herrero J, Garin I, Jugo BM. Diversity and evolution of the Mhc-DRB1 gene in the two endemic Iberian subspecies of Pyrenean chamois, Rupicapra pyrenaica. Heredity. 2007 Oct;99(4):406. - PubMed
-
- Amills M, Jiménez N, Jordana J, Riccardi A, Fernandez-Arias A, Guiral J, Bouzat JL, Folch J, Sanchez A. Low diversity in the major histocompatibility complex class II DRB1 gene of the Spanish ibex, Capra pyrenaica. Heredity. 2004;93(3):266. - PubMed
-
- Miyasaka T, Takeshima SN, Matsumoto Y, Kobayashi N, Matsuhashi T, Miyazaki Y, Tanabe Y, Ishibashi K, Sentsui H, Aida Y. The diversity of bovine MHC class II DRB3 and DQA1 alleles in different herds of Japanese Black and Holstein cattle in Japan. Gene. 2011;472(1):42–9. - PubMed
-
- Takeshima SN, Aida Y. Structure, function and disease susceptibility of the bovine major histocompatibility complex. Anim Sci J. 2006;77(2):138–50.
LinkOut - more resources
Full Text Sources
Other Literature Sources

