An NFκB-dependent mechanism of tumor cell plasticity and lateral transmission of aggressive features
- PMID: 29928478
- PMCID: PMC6003573
- DOI: 10.18632/oncotarget.25465
An NFκB-dependent mechanism of tumor cell plasticity and lateral transmission of aggressive features
Abstract
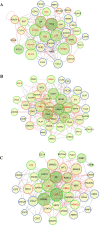
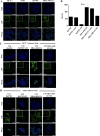
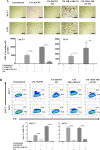
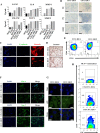
Breast cancer is a complex disease exhibiting extensive inter- and intra-tumor heterogeneity. Inflammation is a well-known driver of cancer progression, often attributed to immune cells infiltrating the tumor stroma. However, tumor cells themselves are capable to secrete a variety of inflammatory molecules, of which we understand very little about their role in intra-clonal communication. We recently reported the capacity of triple negative cell lines to induce a cancer stem cell (CSC)-like phenotype and invasion properties into luminal cells, a mechanism mediated by pro-inflammatory cytokines that up-regulated the CXCL12/CXCR4/CXCR7 chemokine signaling axis. We performed transcriptional array analyses of CSCs-associated genes and cancer-inflammatory cell crosstalk genes and built regulatory networks with the data collected. We found a specific molecular signature segregating with the induced-invasive/stemness phenotype. Regulatory network analysis pointed out to an NFκB transcriptional signature, active in aggressive triple negative cells and in induced-invasive/CSC-like luminal cells. In agreement, NFκB inhibition abolished the induction of the stemness/invasive features. These data support an NFκB dependent mechanism of intra-clonal communication responsible for tumor cell plasticity leading the acquisition of cancer aggressive features. Understanding the communication between different tumor clones would help to find better therapeutic and prophylactic targets to prevent BrC progression and relapse.
Keywords: Intra-tumor heterogeneity and intra-clonal communication; NFκB and STAT signaling pathways; cancer stem cells; invasion; tumor aggression.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare that they have no conflicts of interest.
Figures




References
-
- Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133–45. https://doi.org/10.1002/ijc.27711. - DOI - PubMed
-
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. https://doi.org/10.1002/ijc.29210. - DOI - PubMed
-
- Vargo-Gogola T, Rosen JM. Modelling breast cancer: one size does not fit all. Nat Rev Cancer. 2007;7:659–72. https://doi.org/10.1038/nrc2193. - DOI - PubMed
-
- van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002:415. https://doi.org/10.1038/415530a. - DOI - PubMed
-
- Kwa M, Makris A, Esteva FJ. Clinical utility of gene-expression signatures in early stage breast cancer. Nat Rev Clin Oncol. 2017;14:595. https://doi.org/10.1038/nrclinonc.2017.74. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

