SRSF3 maintains transcriptome integrity in oocytes by regulation of alternative splicing and transposable elements
- PMID: 29928511
- PMCID: PMC6006335
- DOI: 10.1038/s41421-018-0032-3
SRSF3 maintains transcriptome integrity in oocytes by regulation of alternative splicing and transposable elements
Abstract
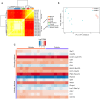
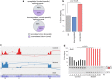
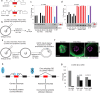
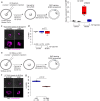
The RNA-binding protein SRSF3 (also known as SRp20) has critical roles in the regulation of pre-mRNA splicing. Zygotic knockout of Srsf3 results in embryo arrest at the blastocyst stage. However, SRSF3 is also present in oocytes, suggesting that it might be critical as a maternally inherited factor. Here we identify SRSF3 as an essential regulator of alternative splicing and of transposable elements to maintain transcriptome integrity in mouse oocyte. Using 3D time-lapse confocal live imaging, we show that conditional deletion of Srsf3 in fully grown germinal vesicle oocytes substantially compromises the capacity of germinal vesicle breakdown (GVBD), and consequently entry into meiosis. By combining single cell RNA-seq, and oocyte micromanipulation with steric blocking antisense oligonucleotides and RNAse-H inducing gapmers, we found that the GVBD defect in mutant oocytes is due to both aberrant alternative splicing and derepression of B2 SINE transposable elements. Together, our study highlights how control of transcriptional identity of the maternal transcriptome by the RNA-binding protein SRSF3 is essential to the development of fertilized-competent oocytes.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

