Bioinformatic analysis of the fold type I PLP-dependent enzymes reveals determinants of reaction specificity in l-threonine aldolase from Aeromonas jandaei
- PMID: 29928580
- PMCID: PMC5986058
- DOI: 10.1002/2211-5463.12441
Bioinformatic analysis of the fold type I PLP-dependent enzymes reveals determinants of reaction specificity in l-threonine aldolase from Aeromonas jandaei
Abstract
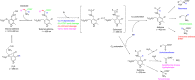
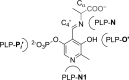
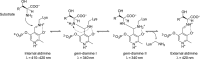
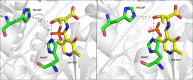
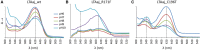
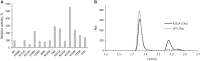
Understanding the role of specific amino acid residues in the molecular mechanism of a protein's function is one of the most challenging problems in modern biology. A systematic bioinformatic analysis of protein families and superfamilies can help in the study of structure-function relationships and in the design of improved variants of enzymes/proteins, but represents a methodological challenge. The pyridoxal-5'-phosphate (PLP)-dependent enzymes are catalytically diverse and include the aspartate aminotransferase superfamily which implements a common structural framework known as type fold I. In this work, the recently developed bioinformatic online methods Mustguseal and Zebra were used to collect and study a large representative set of the aspartate aminotransferase superfamily with high structural, but low sequence similarity to l-threonine aldolase from Aeromonas jandaei (LTAaj), in order to identify conserved positions that provide general properties in the superfamily, and to reveal family-specific positions (FSPs) responsible for functional diversity. The roles of the identified residues in the catalytic mechanism and reaction specificity of LTAaj were then studied by experimental site-directed mutagenesis and molecular modelling. It was shown that FSPs determine reaction specificity by coordinating the PLP cofactor in the enzyme's active centre, thus influencing its activation and the tautomeric equilibrium of the intermediates, which can be used as hotspots to modulate the protein's functional properties. Mutagenesis at the selected FSPs in LTAaj led to a reduction in a native catalytic activity and increased the rate of promiscuous reactions. The results provide insight into the structural basis of catalytic promiscuity of the PLP-dependent enzymes and demonstrate the potential of bioinformatic analysis in studying structure-function relationship in protein superfamilies.
Keywords: bioinformatics; enzyme catalysis; protein engineering; pyridoxal‐phosphate; threonine aldolase.
Figures
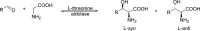




References
-
- Phillips RS (2015) Chemistry and diversity of pyridoxal‐5’‐phosphate dependent enzymes. Biochim Biophys Acta 1854, 1167–1174. - PubMed
-
- Steinreiber J, Fesko K, Reisinger C, Schürmann M, van Assema F, Wolberg M, Mink D and Griengl H (2007) Threonine aldolases ‐ an emerging tool for organic synthesis. Tetrahedron 63, 918–926.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

