Nitric oxide induced S-nitrosation causes base excision repair imbalance
- PMID: 29929044
- PMCID: PMC6436541
- DOI: 10.1016/j.dnarep.2018.04.008
Nitric oxide induced S-nitrosation causes base excision repair imbalance
Abstract
It is well established that inflammation leads to the creation of potent DNA damaging chemicals, including reactive oxygen and nitrogen species. Nitric oxide can react with glutathione to create S-nitrosoglutathione (GSNO), which can in turn lead to S-nitrosated proteins. Of particular interest is the impact of GSNO on the function of DNA repair enzymes. The base excision repair (BER) pathway can be initiated by the alkyl-adenine DNA glycosylase (AAG), a monofunctional glycosylase that removes methylated bases. After base removal, an abasic site is formed, which then gets cleaved by AP endonuclease and processed by downstream BER enzymes. Interestingly, using the Fluorescence-based Multiplexed Host Cell Reactivation Assay (FM-HCR), we show that GSNO actually enhances AAG activity, which is consistent with the literature. This raised the possibility that there might be imbalanced BER when cells are challenged with a methylating agent. To further explore this possibility, we confirmed that GSNO can cause AP endonuclease to translocate from the nucleus to the cytoplasm, which might further exacerbate imbalanced BER by increasing the levels of AP sites. Analysis of abasic sites indeed shows GSNO induces an increase in the level of AP sites. Furthermore, analysis of DNA damage using the CometChip (a higher throughput version of the comet assay) shows an increase in the levels of BER intermediates. Finally, we found that GSNO exposure is associated with an increase in methylation-induced cytotoxicity. Taken together, these studies support a model wherein GSNO increases BER initiation while processing of AP sites is decreased, leading to a toxic increase in BER intermediates. This model is also supported by additional studies performed in our laboratory showing that inflammation in vivo leads to increased large-scale sequence rearrangements. Taken together, this work provides new evidence that inflammatory chemicals can drive cytotoxicity and mutagenesis via BER imbalance.
Keywords: AAG; Base excision repair; DNA alkylation; GSNO; S-Nitrosation.
Copyright © 2018 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


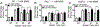

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

