Impact of spliceosome mutations on RNA splicing in myelodysplasia: dysregulated genes/pathways and clinical associations
- PMID: 29930011
- PMCID: PMC6172604
- DOI: 10.1182/blood-2018-04-843771
Impact of spliceosome mutations on RNA splicing in myelodysplasia: dysregulated genes/pathways and clinical associations
Abstract
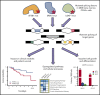
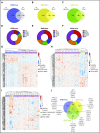
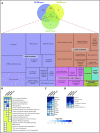
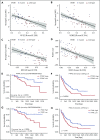
SF3B1, SRSF2, and U2AF1 are the most frequently mutated splicing factor genes in the myelodysplastic syndromes (MDS). We have performed a comprehensive and systematic analysis to determine the effect of these commonly mutated splicing factors on pre-mRNA splicing in the bone marrow stem/progenitor cells and in the erythroid and myeloid precursors in splicing factor mutant MDS. Using RNA-seq, we determined the aberrantly spliced genes and dysregulated pathways in CD34+ cells of 84 patients with MDS. Splicing factor mutations result in different alterations in splicing and largely affect different genes, but these converge in common dysregulated pathways and cellular processes, focused on RNA splicing, protein synthesis, and mitochondrial dysfunction, suggesting common mechanisms of action in MDS. Many of these dysregulated pathways and cellular processes can be linked to the known disease pathophysiology associated with splicing factor mutations in MDS, whereas several others have not been previously associated with MDS, such as sirtuin signaling. We identified aberrantly spliced events associated with clinical variables, and isoforms that independently predict survival in MDS and implicate dysregulation of focal adhesion and extracellular exosomes as drivers of poor survival. Aberrantly spliced genes and dysregulated pathways were identified in the MDS-affected lineages in splicing factor mutant MDS. Functional studies demonstrated that knockdown of the mitosis regulators SEPT2 and AKAP8, aberrantly spliced target genes of SF3B1 and SRSF2 mutations, respectively, led to impaired erythroid cell growth and differentiation. This study illuminates the effect of the common spliceosome mutations on the MDS phenotype and provides novel insights into disease pathophysiology.
© 2018 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures

Comment in
-
An RNA bestiary in splicing-mutant MDS.Blood. 2018 Sep 20;132(12):1217-1219. doi: 10.1182/blood-2018-07-859942. Blood. 2018. PMID: 30237252 No abstract available.
References
-
- Heaney ML, Golde DW. Myelodysplasia. N Engl J Med. 1999;340(21):1649-1660. - PubMed
-
- Pellagatti A, Boultwood J. The molecular pathogenesis of the myelodysplastic syndromes. Eur J Haematol. 2015;95(1):3-15. - PubMed
-
- Tefferi A, Vardiman JW. Myelodysplastic syndromes. N Engl J Med. 2009;361(19):1872-1885. - PubMed
-
- Woll PS, Kjällquist U, Chowdhury O, et al. . Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo [published correction appears in Cancer Cell. 2014;25(6):861]. Cancer Cell. 2014;25(6):794-808. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

