Coactivator condensation at super-enhancers links phase separation and gene control
- PMID: 29930091
- PMCID: PMC6092193
- DOI: 10.1126/science.aar3958
Coactivator condensation at super-enhancers links phase separation and gene control
Abstract
Super-enhancers (SEs) are clusters of enhancers that cooperatively assemble a high density of the transcriptional apparatus to drive robust expression of genes with prominent roles in cell identity. Here we demonstrate that the SE-enriched transcriptional coactivators BRD4 and MED1 form nuclear puncta at SEs that exhibit properties of liquid-like condensates and are disrupted by chemicals that perturb condensates. The intrinsically disordered regions (IDRs) of BRD4 and MED1 can form phase-separated droplets, and MED1-IDR droplets can compartmentalize and concentrate the transcription apparatus from nuclear extracts. These results support the idea that coactivators form phase-separated condensates at SEs that compartmentalize and concentrate the transcription apparatus, suggest a role for coactivator IDRs in this process, and offer insights into mechanisms involved in the control of key cell-identity genes.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures

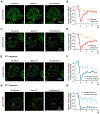




Comment in
-
Concentrating on intrinsic disorder.Nat Rev Mol Cell Biol. 2018 Sep;19(9):544. doi: 10.1038/s41580-018-0039-5. Nat Rev Mol Cell Biol. 2018. PMID: 29993031 No abstract available.
-
Concentrating on intrinsic disorder.Nat Rev Genet. 2018 Sep;19(9):534. doi: 10.1038/s41576-018-0037-7. Nat Rev Genet. 2018. PMID: 30013225 No abstract available.
-
Dynamic condensates activate transcription.Science. 2018 Jul 27;361(6400):329-330. doi: 10.1126/science.aau4795. Science. 2018. PMID: 30049863 No abstract available.
References
-
- Hyman AA, Weber CA, Jülicher F, Liquid-Liquid Phase Separation in Biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014). - PubMed
-
- Shin Y, Brangwynne CP, Liquid phase condensation in cell physiology and disease. Science. 357, eaaf4382 (2017). - PubMed
-
- Brangwynne CP et al. , Germline P Granules Are Liquid Droplets That Localize by Controlled Dissolution/Condensation. Science. 324, 1729–1732 (2009). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

