An Assembly Factor Promotes Assembly of Flavinated SDH1 into the Succinate Dehydrogenase Complex
- PMID: 29930107
- PMCID: PMC6084658
- DOI: 10.1104/pp.18.00320
An Assembly Factor Promotes Assembly of Flavinated SDH1 into the Succinate Dehydrogenase Complex
Abstract
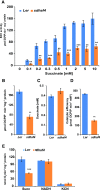
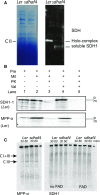
Succinate dehydrogenase (Complex II; SDH) plays an important role in mitochondrial respiratory metabolism. The SDH complex consists of four core subunits and multiple cofactors, which must be assembled correctly to ensure enzyme function. To date, only an assembly factor (SDHAF2) required for FAD insertion into subunit SDH1 has been identified in plants. Here, we report the identification of Arabidopsis (Arabidopsis thaliana) At5g67490 as a second SDH assembly factor. Knockout of At5g67490 (sdhaf4) did not cause any phenotypic variation in seedlings but resulted in a decrease in both SDH activity and the succinate-dependent respiration rate as well as increased accumulation of succinate. Mass spectrometry analyses revealed stable levels of FAD-SDH1 in sdhaf4, together with increased levels of the FAD-SDH1 assembly factor, SDHAF2, and reduced levels of SDH2 compared with the wild type. Loss of SDHAF4 in sdhaf4 inhibited the formation of the SDH1/SDH2 intermediate, leading to the accumulation of soluble SDH1 in the mitochondrial matrix and reduced levels of SDH1 in the membrane. The increased levels of SDHAF2 suggest that the stabilization of soluble FAD-SDH1 depends on SDHAF2 availability. We conclude that SDHAF4 acts on FAD-SDH1 and promotes its assembly with SDH2, thereby stabilizing SDH2 and enabling its full assembly with SDH3/SDH4 to form the SDH complex.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures





References
-
- Araújo WL, Nunes-Nesi A, Osorio S, Usadel B, Fuentes D, Nagy R, Balbo I, Lehmann M, Studart-Witkowski C, Tohge T, et al. (2011) Antisense inhibition of the iron-sulphur subunit of succinate dehydrogenase enhances photosynthesis and growth in tomato via an organic acid-mediated effect on stomatal aperture. Plant Cell 23: 600–627 - PMC - PubMed
-
- Bafunno V, Giancaspero TA, Brizio C, Bufano D, Passarella S, Boles E, Barile M (2004) Riboflavin uptake and FAD synthesis in Saccharomyces cerevisiae mitochondria: involvement of the Flx1p carrier in FAD export. J Biol Chem 279: 95–102 - PubMed
-
- Carrie C, Murcha MW, Kuehn K, Duncan O, Barthet M, Smith PM, Eubel H, Meyer E, Day DA, Millar AH, et al. (2008) Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett 582: 3073–3079 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

