Naked mole rat cells display more efficient excision repair than mouse cells
- PMID: 29930219
- PMCID: PMC6046242
- DOI: 10.18632/aging.101482
Naked mole rat cells display more efficient excision repair than mouse cells
Abstract
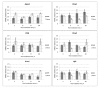
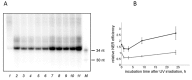
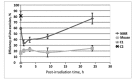
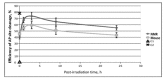
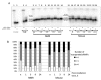
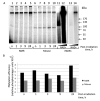
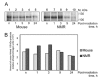
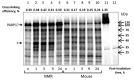
Naked mole rat (NMR) is the long-lived and tumor-resistant rodent. NMRs possess multiple adaptations that may contribute to longevity and cancer-resistance. However, whether NMRs have more efficient DNA repair have not been directly tested. Here we compared base excision repair (BER) and nucleotide excision repair (NER) systems in extracts from NMR and mouse fibroblasts after UVC irradiation. Transcript levels of the key repair enzymes demonstrated in most cases higher inducibility in the mouse vs the NMR cells. Ratios of repair enzymes activities in the extracts somewhat varied depending on post-irradiation time. NMR cell extracts were 2-3-fold more efficient at removing the bulky lesions, 1.5-3-fold more efficient at removing uracil, and about 1.4-fold more efficient at cleaving the AP-site than the mouse cells, while DNA polymerase activities being as a whole higher in the mouse demonstrate different patterns of product distribution. The level of poly(ADP-ribose) synthesis was 1.4-1.8-fold higher in the NMR cells. Furthermore, NMR cell extracts displayed higher binding of PARP1 to DNA probes containing apurinic/apyrimidinic site or photo-reactive DNA lesions. Cumulatively, our results suggest that the NMR has more efficient excision repair systems than the mouse, which may contribute to longevity and cancer resistance of this species.
Keywords: Heterochephalus glaber; Mus musculus; base excision repair; nucleotide excision repair; poly(ADP-ribose) polymerase 1.
Conflict of interest statement
Figures

Similar articles
-
Poly(ADP-ribosyl)ation and DNA repair synthesis in the extracts of naked mole rat, mouse, and human cells.Aging (Albany NY). 2019 May 13;11(9):2852-2873. doi: 10.18632/aging.101959. Aging (Albany NY). 2019. PMID: 31085801 Free PMC article.
-
Use of qPCR to Evaluate Efficiency of the Bulky DNA Damage Removal in Extracts of Mammalian Cells with Different Maximum Lifespan.Biochemistry (Mosc). 2024 Jul;89(7):1183-1191. doi: 10.1134/S0006297924070022. Biochemistry (Mosc). 2024. PMID: 39218017
-
Comparative analysis of the primary structure and production of recombinant poly(ADP-ribose)polymerase 1 of long-lived Heterocephalus glaber.Vavilovskii Zhurnal Genet Selektsii. 2024 Nov;28(7):688-695. doi: 10.18699/vjgb-24-77. Vavilovskii Zhurnal Genet Selektsii. 2024. PMID: 39722667 Free PMC article.
-
Carcinogenesis resistance in the longest-lived rodent, the naked mole-rat.Cancer Sci. 2022 Dec;113(12):4030-4036. doi: 10.1111/cas.15570. Epub 2022 Sep 20. Cancer Sci. 2022. PMID: 36083242 Free PMC article. Review.
-
Uracil-initiated base excision DNA repair synthesis fidelity in human colon adenocarcinoma LoVo and Escherichia coli cell extracts.Prog Nucleic Acid Res Mol Biol. 2001;68:165-88. doi: 10.1016/s0079-6603(01)68098-x. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554295 Review.
Cited by
-
Establishment of primary and immortalized fibroblasts reveals resistance to cytotoxic agents and loss of necroptosis-inducing ability in long-lived Damaraland mole-rats.Geroscience. 2025 Feb;47(1):1381-1396. doi: 10.1007/s11357-024-01420-9. Epub 2024 Dec 2. Geroscience. 2025. PMID: 39623066 Free PMC article.
-
Nontraditional systems in aging research: an update.Cell Mol Life Sci. 2021 Feb;78(4):1275-1304. doi: 10.1007/s00018-020-03658-w. Epub 2020 Oct 9. Cell Mol Life Sci. 2021. PMID: 33034696 Free PMC article. Review.
-
Convergent evolution of a genomic rearrangement may explain cancer resistance in hystrico- and sciuromorpha rodents.NPJ Aging Mech Dis. 2021 Sep 1;7(1):20. doi: 10.1038/s41514-021-00072-9. NPJ Aging Mech Dis. 2021. PMID: 34471123 Free PMC article.
-
Resistance to chemical carcinogenesis induction via a dampened inflammatory response in naked mole-rats.Commun Biol. 2022 Mar 30;5(1):287. doi: 10.1038/s42003-022-03241-y. Commun Biol. 2022. PMID: 35354912 Free PMC article.
-
Chemically induced carcinogenesis in rodent models of aging: assessing organismal resilience to genotoxic stressors in geroscience research.Geroscience. 2019 Apr;41(2):209-227. doi: 10.1007/s11357-019-00064-4. Epub 2019 Apr 29. Geroscience. 2019. PMID: 31037472 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

