Triage of high-risk HPV-positive women in population-based screening by miRNA expression analysis in cervical scrapes; a feasibility study
- PMID: 29930741
- PMCID: PMC5992707
- DOI: 10.1186/s13148-018-0509-9
Triage of high-risk HPV-positive women in population-based screening by miRNA expression analysis in cervical scrapes; a feasibility study
Abstract
Background: Primary testing for high-risk HPV (hrHPV) is increasingly implemented in cervical cancer screening programs. Many hrHPV-positive women, however, harbor clinically irrelevant infections, demanding additional disease markers to prevent over-referral and over-treatment. Most promising biomarkers reflect molecular events relevant to the disease process that can be measured objectively in small amounts of clinical material, such as miRNAs. We previously identified eight miRNAs with altered expression in cervical precancer and cancer due to either methylation-mediated silencing or chromosomal alterations. In this study, we evaluated the clinical value of these eight miRNAs on cervical scrapes to triage hrHPV-positive women in cervical screening.
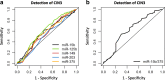
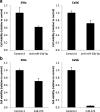
Results: Expression levels of the eight candidate miRNAs in cervical tissue samples (n = 58) and hrHPV-positive cervical scrapes from a screening population (n = 187) and cancer patients (n = 38) were verified by quantitative RT-PCR. In tissue samples, all miRNAs were significantly differentially expressed (p < 0.05) between normal, high-grade precancerous lesions (CIN3), and/or cancer. Expression patterns detected in cervical tissue samples were reflected in cervical scrapes, with five miRNAs showing significantly differential expression between controls and women with CIN3 and cancer. Using logistic regression analysis, a miRNA classifier was built for optimal detection of CIN3 in hrHPV-positive cervical scrapes from the screening population and its performance was evaluated using leave-one-out cross-validation. This miRNA classifier consisted of miR-15b-5p and miR-375 and detected a major subset of CIN3 as well as all carcinomas at a specificity of 70%. The CIN3 detection rate was further improved by combining the two miRNAs with HPV16/18 genotyping. Interestingly, both miRNAs affected the viability of cervical cancer cells in vitro.
Conclusions: This study shows that miRNA expression analysis in cervical scrapes is feasible and enables the early detection of cervical cancer, thus underlining the potential of miRNA expression analysis for triage of hrHPV-positive women in cervical cancer screening.
Keywords: CIN; Cervical cancer; HPV; Scrape; Screening; Triage; miRNA; qRT-PCR.
Conflict of interest statement
This study was approved by the Institutional Review Boards of the VU University Medical Center and Antoni van Leeuwenhoek Hospital. All samples were used in an anonymous fashion in accordance with the “Code for Proper Secondary Use of Human Tissues in the Netherlands” as formulated by the Dutch Federation of Medical Scientific Organizations (https://www.federa.org) [47].DAMH occasionally serves on the scientific advisory board of Pfizer and Bristol-Meyer Squibb and has been on the speakers’ bureau of Qiagen. PJFS, RDMS, CJLMM, and DAMH are minority stakeholders of Self-screen B.V., a spin-off company of VU University Medical Center; and since September 2017, CJLMM is director of Self-screen B.V., which holds patents related to the work. PJFS has received speakers’ bureau honoraria from Roche, Qiagen, Gen-Probe, Abbott, and Seegene. He is consultant for Crucell Holland B.V. CJLMM has participated in the sponsored speaker’s bureau of Merck and Qiagen and served occasionally on the scientific advisory board of Qiagen and Merck. CJLMM owns a small number of shares of Qiagen, has occasionally been consultant for Qiagen, and until April 2016 was a minority shareholder of Diassay B.V. No potential conflicts of interest were disclosed by the other authors.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

