Potential candidate treatment agents for targeting of cholangiocarcinoma identified by gene expression profile analysis
- PMID: 29930804
- PMCID: PMC6007048
- DOI: 10.3892/br.2018.1101
Potential candidate treatment agents for targeting of cholangiocarcinoma identified by gene expression profile analysis
Abstract
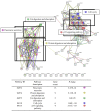
Cholangiocarcinoma (CCA) remains to be a major health problem in several Asian countries including Thailand. The molecular mechanism of CCA is poorly understood. Early diagnosis is difficult, and at present, no effective therapeutic drug is available. The present study aimed to identify the molecular mechanism of CCA by gene expression profile analysis and to search for current approved drugs which may interact with the upregulated genes in CCA. Gene Expression Omnibus (GEO) was used to analyze the gene expression profiles of CCA patients and normal subjects. Using the Kyoto Encyclopedia of Genes and Genomes (KEGG), gene ontology enrichment analysis was also performed, with the KEGG pathway analysis indicating that pancreatic secretion, protein digestion and absorption, fat digestion and absorption, and glycerolipid metabolism may serve important roles in CCA oncogenesis. The drug signature database (DsigDB) was used to search for US Food and Drug Administration (FDA)-approved drugs potentially capable of reversing the effects of the upregulated gene expression in CCA. A total of 61 antineoplastic and 86 non-antineoplastic drugs were identified. Checkpoint kinase 1 was the most interacting with drug signatures. Many of the targeted protein inhibitors that were identified have been approved by the US-FDA as therapeutic agents for non-antineoplastic diseases, including cimetidine, valproic acid and lovastatin. The current study demonstrated an application for bioinformatics analysis in assessing the potential efficacy of currently approved drugs for novel use. The present results suggest novel indications regarding existing drugs useful for CCA treatment. However, further in vitro and in vivo studies are required to support the current predictions.
Keywords: cholangiocarcinoma; gene expression analysis; gene-drug interaction; non-antineoplastic drug; pathway analysis.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
