BioStructMap: a Python tool for integration of protein structure and sequence-based features
- PMID: 29931276
- PMCID: PMC6223362
- DOI: 10.1093/bioinformatics/bty474
BioStructMap: a Python tool for integration of protein structure and sequence-based features
Abstract
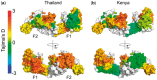
Summary: A sliding window analysis over a protein or genomic sequence is commonly performed, and we present a Python tool, BioStructMap, that extends this concept to three-dimensional (3D) space, allowing the application of a 3D sliding window analysis over a protein structure. BioStructMap is easily extensible, allowing the user to apply custom functions to spatially aggregated data. BioStructMap also allows mapping of underlying genomic sequences to protein structures, allowing the user to perform genetic-based analysis over spatially linked codons-this has applications when selection pressures arise at the level of protein structure.
Availability and implementation: The Python BioStructMap package is available at https://github.com/andrewguy/biostructmap and released under the MIT License. An online server implementing standard functionality is available at https://biostructmap.burnet.edu.au.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

