Asymmetric vasopressin signaling spatially organizes the master circadian clock
- PMID: 29931690
- PMCID: PMC6158041
- DOI: 10.1002/cne.24478
Asymmetric vasopressin signaling spatially organizes the master circadian clock
Abstract
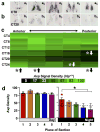
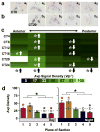
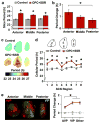
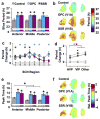
The suprachiasmatic nucleus (SCN) is the neural network that drives daily rhythms in behavior and physiology. The SCN encodes environmental changes through the phasing of cellular rhythms across its anteroposterior axis, but it remains unknown what signaling mechanisms regulate clock function along this axis. Here we demonstrate that arginine vasopressin (AVP) signaling organizes the SCN into distinct anteroposterior domains. Spatial mapping of SCN gene expression using in situ hybridization delineated anterior and posterior domains for AVP signaling components, including complementary patterns of V1a and V1b expression that suggest different roles for these two AVP receptors. Similarly, anteroposterior patterning of transcripts involved in Vasoactive Intestinal Polypeptide- and Prokineticin2 signaling was evident across the SCN. Using bioluminescence imaging, we then revealed that inhibiting V1A and V1B signaling alters period and phase differentially along the anteroposterior SCN. V1 antagonism lengthened period the most in the anterior SCN, whereas changes in phase were largest in the posterior SCN. Further, separately antagonizing V1A and V1B signaling modulated SCN function in a manner that mapped onto anteroposterior expression patterns. Lastly, V1 antagonism influenced SCN period and phase along the dorsoventral axis, complementing effects on the anteroposterior axis. Together, these results indicate that AVP signaling modulates SCN period and phase in a spatially specific manner, which is expected to influence how the master clock interacts with downstream tissues and responds to environmental changes. More generally, we reveal anteroposterior asymmetry in neuropeptide signaling as a recurrent organizational motif that likely influences neural computations in the SCN clock network.
Keywords: RRID: AB_2313978; RRID: AB_2340474; RRID: AB_2556546; RRID: AB_518682; RRID: IMSR_JAX:006852; anteroposterior; circadian; spatiotemporal; suprachiasmatic; vasopressin.
© 2018 Wiley Periodicals, Inc.
Figures









References
-
- Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: Retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916(1–2):172–191. - PubMed
-
- Aschoff J. Response curves in circadian periodicity. In: Aschoff J, editor. Circadian Clocks. Amsterdam: North Holland; 1965. pp. 95–111.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

