Proximal Pathway Enrichment Analysis for Targeting Comorbid Diseases via Network Endopharmacology
- PMID: 29932108
- PMCID: PMC6160959
- DOI: 10.3390/ph11030061
Proximal Pathway Enrichment Analysis for Targeting Comorbid Diseases via Network Endopharmacology
Abstract
The past decades have witnessed a paradigm shift from the traditional drug discovery shaped around the idea of “one target, one disease” to polypharmacology (multiple targets, one disease). Given the lack of clear-cut boundaries across disease (endo)phenotypes and genetic heterogeneity across patients, a natural extension to the current polypharmacology paradigm is to target common biological pathways involved in diseases via endopharmacology (multiple targets, multiple diseases). In this study, we present proximal pathway enrichment analysis (PxEA) for pinpointing drugs that target common disease pathways towards network endopharmacology. PxEA uses the topology information of the network of interactions between disease genes, pathway genes, drug targets and other proteins to rank drugs by their interactome-based proximity to pathways shared across multiple diseases, providing unprecedented drug repurposing opportunities. Using PxEA, we show that many drugs indicated for autoimmune disorders are not necessarily specific to the condition of interest, but rather target the common biological pathways across these diseases. Finally, we provide high scoring drug repurposing candidates that can target common mechanisms involved in type 2 diabetes and Alzheimer’s disease, two conditions that have recently gained attention due to the increased comorbidity among patients.
Keywords: Alzheimer’s disease; autoimmune disorders; comorbidity; drug repurposing; network endopharmacology; proximal pathway enrichment analysis; systems medicine; type 2 diabetes.
Conflict of interest statement
E.G. has received compensation from Scipher Medicine for scientific consulting. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures
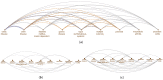
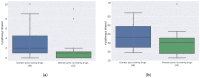

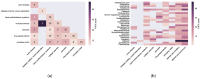
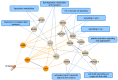
Similar articles
-
Drugs Polypharmacology by In Silico Methods: New Opportunities in Drug Discovery.Curr Pharm Des. 2016;22(21):3073-81. doi: 10.2174/1381612822666160224142323. Curr Pharm Des. 2016. PMID: 26907944 Review.
-
Topology Driven Analysis of Protein - Protein Interactome for Prioritizing Key Comorbid Genes via Sub Graph Based Average Path Length Centrality.IEEE/ACM Trans Comput Biol Bioinform. 2023 Jan-Feb;20(1):742-751. doi: 10.1109/TCBB.2022.3140388. Epub 2023 Feb 3. IEEE/ACM Trans Comput Biol Bioinform. 2023. PMID: 34986099
-
Exploring Polypharmacology in Drug Discovery and Repurposing Using the CANDO Platform.Curr Pharm Des. 2016;22(21):3109-23. doi: 10.2174/1381612822666160325121943. Curr Pharm Des. 2016. PMID: 27013226 Free PMC article. Review.
-
Polypharmacology in Drug Discovery: A Review from Systems Pharmacology Perspective.Curr Pharm Des. 2016;22(21):3171-81. doi: 10.2174/1381612822666160224142812. Curr Pharm Des. 2016. PMID: 26907941 Review.
-
Bioinformatics and network pharmacology analysis of drug targets and mechanisms related to the comorbidity of epilepsy and migraine.Epilepsy Res. 2023 Jan;189:107066. doi: 10.1016/j.eplepsyres.2022.107066. Epub 2022 Dec 15. Epilepsy Res. 2023. PMID: 36571905
Cited by
-
In-silico drug repurposing study predicts the combination of pirfenidone and melatonin as a promising candidate therapy to reduce SARS-CoV-2 infection progression and respiratory distress caused by cytokine storm.PLoS One. 2020 Oct 2;15(10):e0240149. doi: 10.1371/journal.pone.0240149. eCollection 2020. PLoS One. 2020. PMID: 33006999 Free PMC article.
-
Network-based machine learning in colorectal and bladder organoid models predicts anti-cancer drug efficacy in patients.Nat Commun. 2020 Oct 30;11(1):5485. doi: 10.1038/s41467-020-19313-8. Nat Commun. 2020. PMID: 33127883 Free PMC article.
-
A network paradigm predicts drug synergistic effects using downstream protein-protein interactions.CPT Pharmacometrics Syst Pharmacol. 2022 Nov;11(11):1527-1538. doi: 10.1002/psp4.12861. Epub 2022 Oct 6. CPT Pharmacometrics Syst Pharmacol. 2022. PMID: 36204824 Free PMC article.
-
Understanding allergic multimorbidity within the non-eosinophilic interactome.PLoS One. 2019 Nov 6;14(11):e0224448. doi: 10.1371/journal.pone.0224448. eCollection 2019. PLoS One. 2019. PMID: 31693680 Free PMC article.
-
Network Medicine Approach for Analysis of Alzheimer's Disease Gene Expression Data.Int J Mol Sci. 2020 Jan 3;21(1):332. doi: 10.3390/ijms21010332. Int J Mol Sci. 2020. PMID: 31947790 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

