Quantitative Crotonylome Analysis Expands the Roles of p300 in the Regulation of Lysine Crotonylation Pathway
- PMID: 29932303
- PMCID: PMC6420807
- DOI: 10.1002/pmic.201700230
Quantitative Crotonylome Analysis Expands the Roles of p300 in the Regulation of Lysine Crotonylation Pathway
Abstract
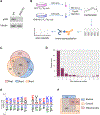
Lysine crotonylation (Kcr) is a recently identified post-translational modification (PTM) that is regulated by an acetyltransferase, p300. The p300-catalyzed histone Kcr is able to stimulate transcription to a greater degree than the well-studied histone lysine acetylation (Kac). Despite these progresses, the global Kcr substrates regulated by p300 remain largely unknown, hindering efforts to establish mechanistic links between Kcr and p300-mediated phenotypes. Here, a quantitative proteomics study to characterize the p300-regulated lysine crotonylome is reported. A total of 816 unique endogenous crotonylation sites are identified across 392 proteins, with 88 sites from 69 proteins being decreased by more than 0.7-fold (log2 < 0.5) and 31 sites from 17 proteins being increased by more than 1.4-fold (log2 > 0.5) in response to p300 knockout (KO). The most downregulated crotonylome alterations under p300 deficiency concern components of the nonsense-mediated decay, infectious disease, and viral/eukaryotic translation pathways. Moreover, some p300-targeted Kcr substrates are potentially linked to diseases such as cancer. Taken together, this study reveals the lysine crotonylome in response to p300, which sheds light on the role for lysine crotonylation in regulation of diverse cellular processes and provides new insights into mechanisms of p300 functions.
Keywords: acetyltransferase; lysine crotonylation; p300; post-translational modification; quantitative proteomics.
© 2018 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
Conflict of Interest
Y.Z. is on the science advisory board of PTM Biolabs. The authors declare no other competing interests.
Figures
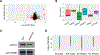


Similar articles
-
Ultradeep Lysine Crotonylome Reveals the Crotonylation Enhancement on Both Histones and Nonhistone Proteins by SAHA Treatment.J Proteome Res. 2017 Oct 6;16(10):3664-3671. doi: 10.1021/acs.jproteome.7b00380. Epub 2017 Sep 19. J Proteome Res. 2017. PMID: 28882038
-
Global Involvement of Lysine Crotonylation in Protein Modification and Transcription Regulation in Rice.Mol Cell Proteomics. 2018 Oct;17(10):1922-1936. doi: 10.1074/mcp.RA118.000640. Epub 2018 Jul 18. Mol Cell Proteomics. 2018. PMID: 30021883 Free PMC article.
-
Cracking Lysine Crotonylation (Kcr): Enlightening a Promising Post-Translational Modification.Chembiochem. 2025 Jan 14;26(2):e202400639. doi: 10.1002/cbic.202400639. Epub 2024 Oct 27. Chembiochem. 2025. PMID: 39462860 Free PMC article. Review.
-
Global crotonylome reveals hypoxia-mediated lamin A crotonylation regulated by HDAC6 in liver cancer.Cell Death Dis. 2022 Aug 17;13(8):717. doi: 10.1038/s41419-022-05165-1. Cell Death Dis. 2022. PMID: 35977926 Free PMC article.
-
Functions and mechanisms of lysine crotonylation.J Cell Mol Med. 2019 Nov;23(11):7163-7169. doi: 10.1111/jcmm.14650. Epub 2019 Sep 1. J Cell Mol Med. 2019. PMID: 31475443 Free PMC article. Review.
Cited by
-
Histone post-translational modification and the DNA damage response.Genes Dis. 2022 Apr 22;10(4):1429-1444. doi: 10.1016/j.gendis.2022.04.002. eCollection 2023 Jul. Genes Dis. 2022. PMID: 37397521 Free PMC article. Review.
-
Function and mechanism of histone β-hydroxybutyrylation in health and disease.Front Immunol. 2022 Sep 12;13:981285. doi: 10.3389/fimmu.2022.981285. eCollection 2022. Front Immunol. 2022. PMID: 36172354 Free PMC article. Review.
-
Lysine crotonylation of DgTIL1 at K72 modulates cold tolerance by enhancing DgnsLTP stability in chrysanthemum.Plant Biotechnol J. 2021 Jun;19(6):1125-1140. doi: 10.1111/pbi.13533. Epub 2021 Jan 21. Plant Biotechnol J. 2021. PMID: 33368971 Free PMC article.
-
Proteomics analysis of lysine crotonylation and 2-hydroxyisobutyrylation reveals significant features of systemic lupus erythematosus.Clin Rheumatol. 2022 Dec;41(12):3851-3858. doi: 10.1007/s10067-022-06254-4. Epub 2022 Aug 8. Clin Rheumatol. 2022. PMID: 35941338 Free PMC article.
-
Qualitative lysine crotonylome analysis in the ovarian tissue of Harmonia axyridis (Pallas).PLoS One. 2021 Oct 18;16(10):e0258371. doi: 10.1371/journal.pone.0258371. eCollection 2021. PLoS One. 2021. PMID: 34662345 Free PMC article.
References
-
- Tremaroli V, Backhed F, Nature 2012, 489, 242. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

