mRNA Deadenylation Is Coupled to Translation Rates by the Differential Activities of Ccr4-Not Nucleases
- PMID: 29932902
- PMCID: PMC6024076
- DOI: 10.1016/j.molcel.2018.05.033
mRNA Deadenylation Is Coupled to Translation Rates by the Differential Activities of Ccr4-Not Nucleases
Abstract
Translation and decay of eukaryotic mRNAs is controlled by shortening of the poly(A) tail and release of the poly(A)-binding protein Pab1/PABP. The Ccr4-Not complex contains two exonucleases-Ccr4 and Caf1/Pop2-that mediate mRNA deadenylation. Here, using a fully reconstituted biochemical system with proteins from the fission yeast Schizosaccharomyces pombe, we show that Pab1 interacts with Ccr4-Not, stimulates deadenylation, and differentiates the roles of the nuclease enzymes. Surprisingly, Pab1 release relies on Ccr4 activity. In agreement with this, in vivo experiments in budding yeast show that Ccr4 is a general deadenylase that acts on all mRNAs. In contrast, Caf1 only trims poly(A) not bound by Pab1. As a consequence, Caf1 is a specialized deadenylase required for the selective deadenylation of transcripts with lower rates of translation elongation and reduced Pab1 occupancy. These findings reveal a coupling between the rates of translation and deadenylation that is dependent on Pab1 and Ccr4-Not.
Keywords: RNA binding protein; deadenylation; exonuclease; gene expression; mRNA decay; poly(A) tail; translation.
Copyright © 2018 MRC Laboratory of Molecular Biology. Published by Elsevier Inc. All rights reserved.
Figures



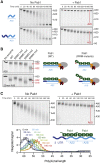
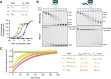


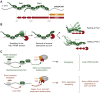
Comment in
-
Tailing Off: PABP and CNOT Generate Cycles of mRNA Deadenylation.Mol Cell. 2018 Jun 21;70(6):987-988. doi: 10.1016/j.molcel.2018.06.009. Mol Cell. 2018. PMID: 29932908
References
-
- Baejen C., Torkler P., Gressel S., Essig K., Söding J., Cramer P. Transcriptome maps of mRNP biogenesis factors define pre-mRNA recognition. Mol. Cell. 2014;55:745–757. - PubMed
-
- Balagopal V., Bolisetty M., Al Husaini N., Coller J., Graveley B.R. Ccr4 and Pop2 control poly(A) tail length in Saccharomyces cerevisiae. bioRxiv. 2017
-
- Beelman C.A., Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J. Biol. Chem. 1994;269:9687–9692. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

