Innate Genetic Evolution of Lung Cancers and Spatial Heterogeneity: Analysis of Treatment-Naïve Lesions
- PMID: 29933065
- PMCID: PMC6153034
- DOI: 10.1016/j.jtho.2018.05.039
Innate Genetic Evolution of Lung Cancers and Spatial Heterogeneity: Analysis of Treatment-Naïve Lesions
Abstract
Introduction: Data regarding the pre-treatment intertumor heterogeneity of potential biomarkers in advanced-stage lung cancers is limited. A finding of such heterogeneity between primary and metastatic lesions would prove valuable to determine if a metastatic lesion can be a surrogate for the primary tumor, as more biomarkers will likely be used in the future to inform treatment decisions.
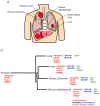
Methods: We performed RNA sequencing to analyze intertumor heterogeneity in 30 specimens (primary tumors, intrathoracic, and extrathoracic metastatic lesions) obtained from five treatment-naïve lung cancer patients.
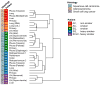
Results: The global unsupervised clustering analysis showed that the lesions clustered at the individual patient level rather than on the metastatic sites, suggesting that the characteristics of specific tumor cells have a greater impact on the gene expression signature than the microenvironment in which the metastasis develops. The mutational and transcriptional data highlight the presence of intertumor heterogeneity showing that the primary tumors are usually distinct from metastatic lesions. Through a comparison between metastatic lesions and the primary tumors, we observed that pathways related to cell proliferation were upregulated, whereas immune-related pathways were downregulated in metastatic lesions.
Conclusion: These data not only provide insight into the evolution of lung cancers, but also imply possibilities and limitations of biomarker-based treatment in lung cancers.
Keywords: Autopsy; Biomarkers; Immune-related markers; RET fusion; RNA sequencing; Tumor heterogeneity.
Copyright © 2018 International Association for the Study of Lung Cancer. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
T. Mitsudomi has received honoraria from AstraZeneca, Chugai, Boehringer-Ingelheim, Pfizer and Roche; has received compensation from AstraZeneca, Chugai, Boehringer-Ingelheim, Pfizer, Roche and Clovis Oncology for participating in advisory boards; and has received research funding (through Kindai University Faculty of Medicine) from AstraZeneca and Chugai. F. R. Hirsch has received compensation from Genentech/Roche, Pfizer, BMS, Lilly, Merck, Ventana/Roche, Novartis and Abbvie for participating in advisory boards; and has received research funding (through the University of Colorado) from Genetech/Roche, BMS, Lilly, Bayer, Amgen and Ventana/Roche. All other authors declare that they have no conflict of interest related to this study.
Our study was supported by an IASLC Young investigator Award (2015 – 2017) to K. Suda, JSPS KAKENHI Grant Number 18K07336 to K. Suda, the Pia and Fred Hirsch Chair in Lung Cancer at the University of Colorado Anschutz Medical Campus, the National Institutes of Health P50CA058187, P30CA046934, Cancer League of Colorado, and the David F. and Margaret T. Grohne Family Foundation.
Figures




References
-
- Hirsch FR, Suda K, Wiens J, Bunn PA., Jr New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet. 2016;388(10048):1012–24. - PubMed
-
- Takano T, Fukui T, Ohe Y, et al. EGFR mutations predict survival benefit from gefitinib in patients with advanced lung adenocarcinoma: a historical comparison of patients treated before and after gefitinib approval in Japan. J Clin Oncol. 2008;26(34):5589–95. - PubMed
-
- Yatabe Y, Matsuo K, Mitsudomi T. Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. J Clin Oncol. 2011;29(22):2972–7. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

