A protein chimera strategy supports production of a model "difficult-to-express" recombinant target
- PMID: 29933498
- PMCID: PMC6174982
- DOI: 10.1002/1873-3468.13170
A protein chimera strategy supports production of a model "difficult-to-express" recombinant target
Abstract
Due in part to the needs of the biopharmaceutical industry, there has been an increased drive to generate high quality recombinant proteins in large amounts. However, achieving high yields can be a challenge as the novelty and increased complexity of new targets often makes them 'difficult-to-express'. This study aimed to define the molecular features that restrict the production of a model 'difficult-to-express' recombinant protein, Tissue Inhibitor Metalloproteinase-3 (TIMP-3). Building from experimental data, computational approaches were used to rationalize the redesign of this recombinant target to generate a chimera with enhanced secretion. The results highlight the importance of early identification of unfavourable sequence attributes, enabling the generation of engineered protein forms that bypass 'secretory' bottlenecks and result in efficient recombinant protein production.
Keywords: difficult-to-express; mammalian expression system; predictive computational tool; protein engineering; recombinant protein production; tissue inhibitor of metalloproteinase.
© 2018 The Authors. FEBS Letters published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Figures

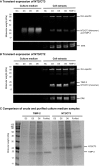
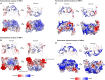
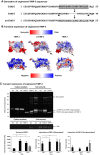
References
-
- Zhu J (2012) Mammalian cell protein expression for biopharmaceutical production. Biotechnol Adv 30, 1158–1170. - PubMed
-
- Walsh G (2014) Biopharmaceutical benchmarks 2014. Nat Biotechnol 32, 992–1000. - PubMed
-
- Hussain H, Maldonado‐Agurto R and Dickson AJ (2014) The endoplasmic reticulum and unfolded protein response in the control of mammalian recombinant protein production. Biotechnol Lett 36, 1581–1593. - PubMed
-
- Fischer S, Handrick R and Otte K (2015) The art of CHO cell engineering: a comprehensive retrospect and future perspectives. Biotechnol Adv 33, 1878–1896. - PubMed
-
- Hansen HG, Pristovšek N, Kildegaard HF and Lee GM (2017) Improving the secretory capacity of Chinese hamster ovary cells by ectopic expression of effector genes: lessons learned and future directions. Biotechnol Adv 35, 64–76. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

