p190 RhoGAP promotes contact inhibition in epithelial cells by repressing YAP activity
- PMID: 29934311
- PMCID: PMC6122998
- DOI: 10.1083/jcb.201710058
p190 RhoGAP promotes contact inhibition in epithelial cells by repressing YAP activity
Erratum in
-
Correction: p190 RhoGAP promotes contact inhibition in epithelial cells by repressing YAP activity.J Cell Biol. 2018 Sep 3;217(9):3313. doi: 10.1083/JCB.20171005807172018c. Epub 2018 Jul 25. J Cell Biol. 2018. PMID: 30045860 Free PMC article. No abstract available.
Abstract
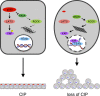
ARHGAP35 encoding p190A RhoGAP is a cancer-associated gene with a mutation spectrum suggestive of a tumor-suppressor function. In this study, we demonstrate that loss of heterozygosity for ARHGAP35 occurs in human tumors. We sought to identify tumor-suppressor capacities for p190A RhoGAP (p190A) and its paralog p190B in epithelial cells. We reveal an essential role for p190A and p190B to promote contact inhibition of cell proliferation (CIP), a function that relies on RhoGAP activity. Unbiased mRNA sequencing analyses establish that p190A and p190B modulate expression of genes associated with the Hippo pathway. Accordingly, we determine that p190A and p190B induce CIP by repressing YAP-TEAD-regulated gene transcription through activation of LATS kinases and inhibition of the Rho-ROCK pathway. Finally, we demonstrate that loss of a single p190 paralog is sufficient to elicit nuclear translocation of YAP and perturb CIP in epithelial cells cultured in Matrigel. Collectively, our data reveal a novel mechanism consistent with a tumor-suppressor function for ARHGAP35.
© 2018 Frank et al.
Figures






Similar articles
-
p120 RasGAP and ZO-2 are essential for Hippo signaling and tumor-suppressor function mediated by p190A RhoGAP.Cell Rep. 2023 Dec 26;42(12):113486. doi: 10.1016/j.celrep.2023.113486. Epub 2023 Nov 22. Cell Rep. 2023. PMID: 37995182 Free PMC article.
-
p190A RhoGAP induces CDH1 expression and cooperates with E-cadherin to activate LATS kinases and suppress tumor cell growth.Oncogene. 2020 Aug;39(33):5570-5587. doi: 10.1038/s41388-020-1385-2. Epub 2020 Jul 8. Oncogene. 2020. PMID: 32641858 Free PMC article.
-
p190B RhoGAP regulates endothelial-cell-associated proteolysis through MT1-MMP and MMP2.J Cell Sci. 2008 Jun 15;121(Pt 12):2054-61. doi: 10.1242/jcs.025817. Epub 2008 May 27. J Cell Sci. 2008. PMID: 18505793
-
Angiomotin'g YAP into the nucleus for cell proliferation and cancer development.Sci Signal. 2013 Sep 3;6(291):pe27. doi: 10.1126/scisignal.2004573. Sci Signal. 2013. PMID: 24003252 Review.
-
Regulation and functions of the DLC family of RhoGAP proteins: Implications for development and cancer.Cell Signal. 2025 Jan;125:111505. doi: 10.1016/j.cellsig.2024.111505. Epub 2024 Nov 15. Cell Signal. 2025. PMID: 39549821 Review.
Cited by
-
Mechanoregulation of YAP and TAZ in Cellular Homeostasis and Disease Progression.Front Cell Dev Biol. 2021 May 24;9:673599. doi: 10.3389/fcell.2021.673599. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34109179 Free PMC article. Review.
-
Genomic Comparison of Endometrioid Endometrial Carcinoma and Its Precancerous Lesions in Chinese Patients by High-Depth Next Generation Sequencing.Front Oncol. 2019 Mar 4;9:123. doi: 10.3389/fonc.2019.00123. eCollection 2019. Front Oncol. 2019. PMID: 30886832 Free PMC article.
-
p120 RasGAP and ZO-2 are essential for Hippo signaling and tumor-suppressor function mediated by p190A RhoGAP.Cell Rep. 2023 Dec 26;42(12):113486. doi: 10.1016/j.celrep.2023.113486. Epub 2023 Nov 22. Cell Rep. 2023. PMID: 37995182 Free PMC article.
-
Fixing the GAP: The role of RhoGAPs in cancer.Eur J Cell Biol. 2022 Apr;101(2):151209. doi: 10.1016/j.ejcb.2022.151209. Epub 2022 Feb 10. Eur J Cell Biol. 2022. PMID: 35180567 Free PMC article. Review.
-
Hippo Signaling in the Endometrium.Int J Mol Sci. 2022 Mar 31;23(7):3852. doi: 10.3390/ijms23073852. Int J Mol Sci. 2022. PMID: 35409214 Free PMC article. Review.
References
-
- Bass A.J., Thorsson V., Shmulevich I., Reynolds S.M., Miller M., Bernard B., Hinoue T., Laird P.W., Curtis C., Shen H., et al. Cancer Genome Atlas Research Network . 2014. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 513:202–209. 10.1038/nature13480 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

