Ethylphenol Formation by Lactobacillus plantarum: Identification of the Enzyme Involved in the Reduction of Vinylphenols
- PMID: 29934329
- PMCID: PMC6102998
- DOI: 10.1128/AEM.01064-18
Ethylphenol Formation by Lactobacillus plantarum: Identification of the Enzyme Involved in the Reduction of Vinylphenols
Abstract
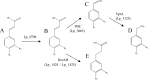
Ethylphenols are strong odorants produced by microbial activity that are described as off flavors in several foods. Lactobacillus plantarum is a lactic acid bacterial species able to produce ethylphenols by the reduction of vinylphenols during the metabolism of hydroxycinnamic acids. However, the reductase involved has not been yet uncovered. In this study, the involvement in vinylphenol reduction of a gene encoding a putative reductase (lp_3125) was confirmed by the absence of reduction activity in the Δlp_3125 knockout mutant. The protein encoded by lp_3125, VprA, was recombinantly produced in Escherichia coli VprA was assayed against vinylphenols (4-vinylphenol, 4-vinylcatechol, and 4-vinylguaiacol), and all were reduced to their corresponding ethylphenols (4-ethylphenol, 4-ethylcatechol, and 4-ethylguaiacol). PCR and high-performance liquid chromatography (HPLC) detection methods revealed that the VprA reductase is not widely distributed among the lactic acid bacteria studied and that only the bacteria possessing the vprA gene were able to produce ethylphenol from vinylphenol. However, all the species belonging to the L. plantarum group were ethylphenol producers. The identification of the L. plantarum VprA protein involved in hydroxycinnamate degradation completes the route of degradation of these compounds in lactic acid bacteria.IMPORTANCE The presence of volatile phenols is considered a major organoleptic defect of several fermented alcoholic beverages. The biosynthesis of these compounds has been mainly associated with Brettanomyces/Dekkera yeasts. However, the potential importance of lactic acid bacteria in volatile phenol spoilage is emphasized by reports describing a faster ethylphenol production by these bacteria than by yeasts. The genetic identification of the bacterial vinylphenol reductase involved in volatile phenol production provides new insights into the role of lactic acid bacteria in the production of these off flavors. The development of a molecular method for the detection of ethylphenol-producing bacteria could be helpful to design strategies to reduce the bacterial production of vinylphenols in fermented foods.
Keywords: aroma; cider; ethylguaiacol; ethylphenol; lactic acid bacteria; off flavors; phenolic compounds; spoilage; volatile phenols; wine.
Copyright © 2018 American Society for Microbiology.
Figures



Similar articles
-
Unravelling the Reduction Pathway as an Alternative Metabolic Route to Hydroxycinnamate Decarboxylation in Lactobacillus plantarum.Appl Environ Microbiol. 2018 Jul 17;84(15):e01123-18. doi: 10.1128/AEM.01123-18. Print 2018 Aug 1. Appl Environ Microbiol. 2018. PMID: 29776925 Free PMC article.
-
Screening of representative cider yeasts and bacteria for volatile phenol-production ability.Food Microbiol. 2011 Oct;28(7):1243-51. doi: 10.1016/j.fm.2011.05.001. Epub 2011 May 19. Food Microbiol. 2011. PMID: 21839372
-
Partial vinylphenol reductase purification and characterization from Brettanomyces bruxellensis.FEMS Microbiol Lett. 2008 Jul;284(2):213-7. doi: 10.1111/j.1574-6968.2008.01192.x. FEMS Microbiol Lett. 2008. PMID: 18576949
-
4-Ethylphenol, 4-ethylguaiacol and 4-ethylcatechol in red wines: Microbial formation, prevention, remediation and overview of analytical approaches.Crit Rev Food Sci Nutr. 2019;59(9):1367-1391. doi: 10.1080/10408398.2017.1408563. Epub 2017 Dec 19. Crit Rev Food Sci Nutr. 2019. PMID: 29257912 Review.
-
Challenges and advances in biotechnological approaches for the synthesis of canolol and other vinylphenols from biobased p-hydroxycinnamic acids: a review.Biotechnol Biofuels Bioprod. 2023 Nov 14;16(1):173. doi: 10.1186/s13068-023-02425-w. Biotechnol Biofuels Bioprod. 2023. PMID: 37964324 Free PMC article. Review.
Cited by
-
Conversion of (poly)phenolic compounds in food fermentations by lactic acid bacteria: Novel insights into metabolic pathways and functional metabolites.Curr Res Food Sci. 2023 Jan 18;6:100448. doi: 10.1016/j.crfs.2023.100448. eCollection 2023. Curr Res Food Sci. 2023. PMID: 36713641 Free PMC article. Review.
-
Microbial regulation of offspring diseases mediated by maternal-associated microbial metabolites.Front Microbiol. 2022 Nov 4;13:955297. doi: 10.3389/fmicb.2022.955297. eCollection 2022. Front Microbiol. 2022. PMID: 36406399 Free PMC article. Review.
-
How Microbiome Composition Correlates with Biochemical Changes during Sauerkraut Fermentation: a Focus on Neglected Bacterial Players and Functionalities.Microbiol Spectr. 2022 Aug 31;10(4):e0016822. doi: 10.1128/spectrum.00168-22. Epub 2022 Jun 14. Microbiol Spectr. 2022. PMID: 35699432 Free PMC article.
-
Biotransformation of Phenolic Acids in Foods: Pathways, Key Enzymes, and Technological Applications.Foods. 2025 Jun 23;14(13):2187. doi: 10.3390/foods14132187. Foods. 2025. PMID: 40646939 Free PMC article. Review.
-
Interaction of 4-ethylphenol, pH, sucrose and ethanol on the growth and fermentation capacity of the industrial strain of Saccharomyces cerevisiae PE-2.World J Microbiol Biotechnol. 2019 Aug 20;35(9):136. doi: 10.1007/s11274-019-2714-x. World J Microbiol Biotechnol. 2019. PMID: 31432249
References
-
- Tempère S, Cuzange E, Schaaper MH, de Lescar R, de Revel G, Sicard G. 2014. “Brett character” in wine? Is there a consensus among professional assessors? A perceptual and conceptual approach. Food Qual Pref 34:29–36.
-
- Chatonnet P, Dubordieu D, Boidron JN, Pons M. 1992. The origin of ethylphenols in wines. J Sci Food Agric 60:165–178. doi:10.1002/jsfa.2740600205. - DOI
-
- Buron N, Coton M, Legendre P, Ledauphin J, Kientz-Bouchart V, Guichard H, Barillier D, Coton E. 2012. Implications of Lactobacillus collinoides and Brettanomyces/Dekkera anomala in phenolic off-flavour defects of ciders. Int J Food Microbiol 153:159–165. doi:10.1016/j.ijfoodmicro.2011.11.002. - DOI - PubMed
-
- Suarez R, Suarez-Lepe JA, Morata A, Calderón F. 2007. The production of ethylphenols in wine by yeasts of the genera Brettanomyces and Dekkera. A review. Food Chem 102:10–21. doi:10.1016/j.foodchem.2006.03.030. - DOI
-
- Shahidi F, Naczk M. 2003. Phenolics in food and nutraceuticals. CRC Press, London, United Kingdom.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

