PTPσ Drives Excitatory Presynaptic Assembly via Various Extracellular and Intracellular Mechanisms
- PMID: 29934346
- PMCID: PMC6705959
- DOI: 10.1523/JNEUROSCI.0672-18.2018
PTPσ Drives Excitatory Presynaptic Assembly via Various Extracellular and Intracellular Mechanisms
Abstract
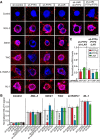
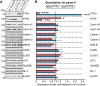
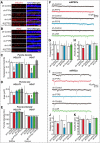
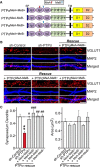
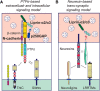
Leukocyte common antigen-receptor protein tyrosine phosphatases (LAR-RPTPs) are hub proteins that organize excitatory and inhibitory synapse development through binding to various extracellular ligands. Here, we report that knockdown (KD) of the LAR-RPTP family member PTPσ reduced excitatory synapse number and transmission in cultured rat hippocampal neurons, whereas KD of PTPδ produced comparable decreases at inhibitory synapses, in both cases without altering expression levels of interacting proteins. An extensive series of rescue experiments revealed that extracellular interactions of PTPσ with Slitrks are important for excitatory synapse development. These experiments further showed that the intracellular D2 domain of PTPσ is required for induction of heterologous synapse formation by Slitrk1 or TrkC, suggesting that interaction of LAR-RPTPs with distinct intracellular presynaptic proteins, drives presynaptic machinery assembly. Consistent with this, double-KD of liprin-α2 and -α3 or KD of PTPσ substrates (N-cadherin and p250RhoGAP) in neurons inhibited Slitrk6-induced, PTPσ-mediated heterologous synapse formation activity. We propose a synaptogenesis model in presynaptic neurons involving LAR-RPTP-organized retrograde signaling cascades, in which both extracellular and intracellular mechanisms are critical in orchestrating distinct synapse types.SIGNIFICANCE STATEMENT In this study, we sought to test the unproven hypothesis that PTPσ and PTPδ are required for excitatory and inhibitory synapse formation/transmission, respectively, in cultured hippocampal neurons, using knockdown-based loss-of-function analyses. We further performed extensive structure-function analyses, focusing on PTPσ-mediated actions, to address the mechanisms of presynaptic assembly at excitatory synaptic sites. Using interdisciplinary approaches, we systematically applied a varied set of PTPσ deletion variants, point mutants, and splice variants to demonstrate that both extracellular and intracellular mechanisms are involved in organizing presynaptic assembly. Strikingly, extracellular interactions of PTPσ with heparan sulfates and Slitrks, intracellular interactions of PTPσ with liprin-α and its associated proteins through the D2 domain, as well as distinct substrates are all critical.
Keywords: LAR-RPTPs; PTPσ; presynaptic assembly; protein–protein interaction; synaptic adhesion molecule.
Copyright © 2018 the authors 0270-6474/18/386700-22$15.00/0.
Figures




Similar articles
-
LAR-RPTPs Directly Interact with Neurexins to Coordinate Bidirectional Assembly of Molecular Machineries.J Neurosci. 2020 Oct 28;40(44):8438-8462. doi: 10.1523/JNEUROSCI.1091-20.2020. Epub 2020 Oct 9. J Neurosci. 2020. PMID: 33037075 Free PMC article.
-
Neurotrophin-3 Regulates Synapse Development by Modulating TrkC-PTPσ Synaptic Adhesion and Intracellular Signaling Pathways.J Neurosci. 2016 Apr 27;36(17):4816-31. doi: 10.1523/JNEUROSCI.4024-15.2016. J Neurosci. 2016. PMID: 27122038 Free PMC article.
-
Emergent Synapse Organizers: LAR-RPTPs and Their Companions.Int Rev Cell Mol Biol. 2016;324:39-65. doi: 10.1016/bs.ircmb.2016.01.002. Epub 2016 Feb 8. Int Rev Cell Mol Biol. 2016. PMID: 27017006 Review.
-
Receptor protein tyrosine phosphatase delta is not essential for synapse maintenance or transmission at hippocampal synapses.Mol Brain. 2020 Jun 17;13(1):94. doi: 10.1186/s13041-020-00629-x. Mol Brain. 2020. PMID: 32552840 Free PMC article.
-
Emerging roles of the neurotrophin receptor TrkC in synapse organization.Neurosci Res. 2017 Mar;116:10-17. doi: 10.1016/j.neures.2016.09.009. Epub 2016 Sep 30. Neurosci Res. 2017. PMID: 27697534 Review.
Cited by
-
Structural insights into selective interaction between type IIa receptor protein tyrosine phosphatases and Liprin-α.Nat Commun. 2020 Jan 31;11(1):649. doi: 10.1038/s41467-020-14516-5. Nat Commun. 2020. PMID: 32005855 Free PMC article.
-
Assembly of the presynaptic active zone.Curr Opin Neurobiol. 2020 Aug;63:95-103. doi: 10.1016/j.conb.2020.03.008. Epub 2020 May 11. Curr Opin Neurobiol. 2020. PMID: 32403081 Free PMC article. Review.
-
PTPσ Knockdown in Lampreys Impairs Reticulospinal Axon Regeneration and Neuronal Survival After Spinal Cord Injury.Front Cell Neurosci. 2020 Mar 19;14:61. doi: 10.3389/fncel.2020.00061. eCollection 2020. Front Cell Neurosci. 2020. PMID: 32265663 Free PMC article.
-
High temporal frequency light response in mouse retina requires FAT3 signaling in bipolar cells.bioRxiv [Preprint]. 2024 Jun 28:2023.11.02.565326. doi: 10.1101/2023.11.02.565326. bioRxiv. 2024. Update in: J Gen Physiol. 2025 Mar 03;157(2):e202413642. doi: 10.1085/jgp.202413642. PMID: 37961274 Free PMC article. Updated. Preprint.
-
Perineuronal Net Receptor PTPσ Regulates Retention of Memories.Front Synaptic Neurosci. 2021 Jul 22;13:672475. doi: 10.3389/fnsyn.2021.672475. eCollection 2021. Front Synaptic Neurosci. 2021. PMID: 34366821 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
