Pulmonary pericytes regulate lung morphogenesis
- PMID: 29934496
- PMCID: PMC6015030
- DOI: 10.1038/s41467-018-04913-2
Pulmonary pericytes regulate lung morphogenesis
Abstract
Blood vessels are essential for blood circulation but also control organ growth, homeostasis, and regeneration, which has been attributed to the release of paracrine signals by endothelial cells. Endothelial tubules are associated with specialised mesenchymal cells, termed pericytes, which help to maintain vessel wall integrity. Here we identify pericytes as regulators of epithelial and endothelial morphogenesis in postnatal lung. Mice lacking expression of the Hippo pathway components YAP and TAZ in pericytes show defective alveologenesis. Mutant pericytes are present in normal numbers but display strongly reduced expression of hepatocyte growth factor leading to impaired activation of the c-Met receptor, which is expressed by alveolar epithelial cells. YAP and TAZ are also required for expression of angiopoietin-1 by pulmonary pericytes, which also controls hepatocyte growth factor expression and thereby alveologenesis in an autocrine fashion. These findings establish that pericytes have important, organ-specific signalling properties and coordinate the behavior of epithelial and vascular cells during lung morphogenesis.
Conflict of interest statement
The authors declare no competing interests.
Figures
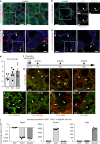
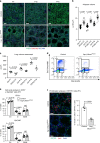


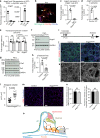
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

