Quantitative assessment of individual populations within polymicrobial biofilms
- PMID: 29934504
- PMCID: PMC6015014
- DOI: 10.1038/s41598-018-27497-9
Quantitative assessment of individual populations within polymicrobial biofilms
Abstract
Selecting appropriate tools providing reliable quantitative measures of individual populations in biofilms is critical as we now recognize their true polymicrobial and heterogeneous nature. Here, plate count, quantitative real-time polymerase chain reaction (q-PCR) and peptide nucleic acid probe-fluorescence in situ hybridization (PNA-FISH) were employed to quantitate cystic fibrosis multispecies biofilms. Growth of Pseudomonas aeruginosa, Inquilinus limosus and Dolosigranulum pigrum was assessed in dual- and triple-species consortia under oxygen and antibiotic stress. Quantification methods, that were previously optimized and validated in planktonic consortia, were not always in agreement when applied in multispecies biofilms. Discrepancies in culture and molecular outcomes were observed, particularly for triple-species consortia and antibiotic-stressed biofilms. Some differences were observed, such as the higher bacterial counts obtained by q-PCR and/or PNA-FISH (≤4 log10 cells/cm2) compared to culture. But the discrepancies between PNA-FISH and q-PCR data (eg D. pigrum limited assessment by q-PCR) demonstrate the effect of biofilm heterogeneity in method's reliability. As the heterogeneity in biofilms is a reflection of a myriad of variables, tailoring an accurate picture of communities´ changes is crucial. This work demonstrates that at least two, but preferentially three, quantification techniques are required to obtain reliable measures and take comprehensive analysis of polymicrobial biofilm-associated infections.
Conflict of interest statement
The authors declare no competing interests.
Figures

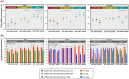
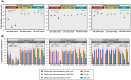

Similar articles
-
Antibiotic resistance of mixed biofilms in cystic fibrosis: impact of emerging microorganisms on treatment of infection.Int J Antimicrob Agents. 2012 Sep;40(3):260-3. doi: 10.1016/j.ijantimicag.2012.04.020. Epub 2012 Jul 4. Int J Antimicrob Agents. 2012. PMID: 22770521
-
Developing a model for cystic fibrosis sociomicrobiology based on antibiotic and environmental stress.Int J Med Microbiol. 2017 Dec;307(8):460-470. doi: 10.1016/j.ijmm.2017.09.018. Epub 2017 Sep 28. Int J Med Microbiol. 2017. PMID: 29033313
-
Alloiococcus otitidis Forms Multispecies Biofilm with Haemophilus influenzae: Effects on Antibiotic Susceptibility and Growth in Adverse Conditions.Front Cell Infect Microbiol. 2017 Aug 2;7:344. doi: 10.3389/fcimb.2017.00344. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28824879 Free PMC article.
-
Pseudomonas aeruginosa Biofilm, a Programmed Bacterial Life for Fitness.J Microbiol Biotechnol. 2017 Jun 28;27(6):1053-1064. doi: 10.4014/jmb.1611.11056. J Microbiol Biotechnol. 2017. PMID: 28301918 Review.
-
Pseudomonas aeruginosa hypoxic or anaerobic biofilm infections within cystic fibrosis airways.Trends Microbiol. 2009 Mar;17(3):130-8. doi: 10.1016/j.tim.2008.12.003. Epub 2009 Feb 21. Trends Microbiol. 2009. PMID: 19231190 Review.
Cited by
-
The Emerging Trend of Bio-Engineering Approaches for Microbial Nanomaterial Synthesis and Its Applications.Front Microbiol. 2021 Mar 16;12:638003. doi: 10.3389/fmicb.2021.638003. eCollection 2021. Front Microbiol. 2021. PMID: 33796089 Free PMC article. Review.
-
An Indirect Fluorescence Microscopy Method to Assess Vaginal Lactobacillus Concentrations.Microorganisms. 2024 Jan 5;12(1):114. doi: 10.3390/microorganisms12010114. Microorganisms. 2024. PMID: 38257941 Free PMC article.
-
A New PNA-FISH Probe Targeting Fannyhessea vaginae.Front Cell Infect Microbiol. 2021 Nov 18;11:779376. doi: 10.3389/fcimb.2021.779376. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34869078 Free PMC article.
-
Influence of the lung microbiome on antibiotic susceptibility of cystic fibrosis pathogens.Eur Respir Rev. 2019 Jul 8;28(152):190041. doi: 10.1183/16000617.0041-2019. Print 2019 Jun 30. Eur Respir Rev. 2019. PMID: 31285289 Free PMC article. Review.
-
RNA-based qPCR as a tool to quantify and to characterize dual-species biofilms.Sci Rep. 2019 Sep 20;9(1):13639. doi: 10.1038/s41598-019-50094-3. Sci Rep. 2019. PMID: 31541147 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

