PTEN-L is a novel protein phosphatase for ubiquitin dephosphorylation to inhibit PINK1-Parkin-mediated mitophagy
- PMID: 29934616
- PMCID: PMC6082900
- DOI: 10.1038/s41422-018-0056-0
PTEN-L is a novel protein phosphatase for ubiquitin dephosphorylation to inhibit PINK1-Parkin-mediated mitophagy
Erratum in
-
Author Correction: PTEN-L is a novel protein phosphatase for ubiquitin dephosphorylation to inhibit PINK1-Parkin-mediated mitophagy.Cell Res. 2018 Aug;28(8):872-873. doi: 10.1038/s41422-018-0068-9. Cell Res. 2018. PMID: 30038297 Free PMC article.
Abstract
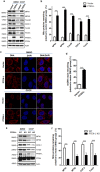
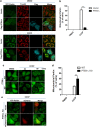
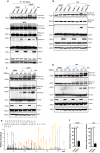
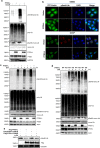
Mitophagy is an important type of selective autophagy for specific elimination of damaged mitochondria. PTEN-induced putative kinase protein 1 (PINK1)-catalyzed phosphorylation of ubiquitin (Ub) plays a critical role in the onset of PINK1-Parkin-mediated mitophagy. Phosphatase and tensin homolog (PTEN)-long (PTEN-L) is a newly identified isoform of PTEN, with addition of 173 amino acids to its N-terminus. Here we report that PTEN-L is a novel negative regulator of mitophagy via its protein phosphatase activity against phosphorylated ubiquitin. We found that PTEN-L localizes at the outer mitochondrial membrane (OMM) and overexpression of PTEN-L inhibits, whereas deletion of PTEN-L promotes, mitophagy induced by various mitochondria-damaging agents. Mechanistically, PTEN-L is capable of effectively preventing Parkin mitochondrial translocation, reducing Parkin phosphorylation, maintaining its closed inactive conformation, and inhibiting its E3 ligase activity. More importantly, PTEN-L reduces the level of phosphorylated ubiquitin (pSer65-Ub) in vivo, and in vitro phosphatase assay confirms that PTEN-L dephosphorylates pSer65-Ub via its protein phosphatase activity, independently of its lipid phosphatase function. Taken together, our findings demonstrate a novel function of PTEN-L as a protein phosphatase for ubiquitin, which counteracts PINK1-mediated ubiquitin phosphorylation leading to blockage of the feedforward mechanisms in mitophagy induction and eventual suppression of mitophagy. Thus, understanding this novel function of PTEN-L provides a key missing piece in the molecular puzzle controlling mitophagy, a critical process in many important human diseases including neurodegenerative disorders such as Parkinson's disease.
Conflict of interest statement
The authors declare no competing interests.
Figures




Comment in
-
PTEN-L puts a brake on mitophagy.Autophagy. 2018;14(11):2023-2025. doi: 10.1080/15548627.2018.1502565. Epub 2018 Sep 2. Autophagy. 2018. PMID: 30106322 Free PMC article.
Similar articles
-
PTEN-L puts a brake on mitophagy.Autophagy. 2018;14(11):2023-2025. doi: 10.1080/15548627.2018.1502565. Epub 2018 Sep 2. Autophagy. 2018. PMID: 30106322 Free PMC article.
-
N-degron-mediated degradation and regulation of mitochondrial PINK1 kinase.Curr Genet. 2020 Aug;66(4):693-701. doi: 10.1007/s00294-020-01062-2. Epub 2020 Mar 10. Curr Genet. 2020. PMID: 32157382 Review.
-
Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy.Proc Natl Acad Sci U S A. 2015 May 26;112(21):6637-42. doi: 10.1073/pnas.1506593112. Epub 2015 May 12. Proc Natl Acad Sci U S A. 2015. PMID: 25969509 Free PMC article.
-
Phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1)-dependent ubiquitination of endogenous Parkin attenuates mitophagy: study in human primary fibroblasts and induced pluripotent stem cell-derived neurons.J Biol Chem. 2013 Jan 25;288(4):2223-37. doi: 10.1074/jbc.M112.391680. Epub 2012 Dec 4. J Biol Chem. 2013. PMID: 23212910 Free PMC article.
-
The three 'P's of mitophagy: PARKIN, PINK1, and post-translational modifications.Genes Dev. 2015 May 15;29(10):989-99. doi: 10.1101/gad.262758.115. Genes Dev. 2015. PMID: 25995186 Free PMC article. Review.
Cited by
-
Simultaneous treatment with sorafenib and glucose restriction inhibits hepatocellular carcinoma in vitro and in vivo by impairing SIAH1-mediated mitophagy.Exp Mol Med. 2022 Nov;54(11):2007-2021. doi: 10.1038/s12276-022-00878-x. Epub 2022 Nov 16. Exp Mol Med. 2022. PMID: 36385558 Free PMC article.
-
p62/SQSTM1 and Selective Autophagy in Cardiometabolic Diseases.Antioxid Redox Signal. 2019 Aug 20;31(6):458-471. doi: 10.1089/ars.2018.7649. Epub 2019 Feb 11. Antioxid Redox Signal. 2019. PMID: 30588824 Free PMC article. Review.
-
Molecular Signaling to Preserve Mitochondrial Integrity against Ischemic Stress in the Heart: Rescue or Remove Mitochondria in Danger.Cells. 2021 Nov 27;10(12):3330. doi: 10.3390/cells10123330. Cells. 2021. PMID: 34943839 Free PMC article. Review.
-
Disruption of Mitochondrial Homeostasis: The Role of PINK1 in Parkinson's Disease.Cells. 2021 Nov 4;10(11):3022. doi: 10.3390/cells10113022. Cells. 2021. PMID: 34831247 Free PMC article. Review.
-
Mitophagy: Molecular Mechanisms, New Concepts on Parkin Activation and the Emerging Role of AMPK/ULK1 Axis.Cells. 2021 Dec 23;11(1):30. doi: 10.3390/cells11010030. Cells. 2021. PMID: 35011593 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

