The A-Z of Zika drug discovery
- PMID: 29935345
- PMCID: PMC7108251
- DOI: 10.1016/j.drudis.2018.06.014
The A-Z of Zika drug discovery
Abstract
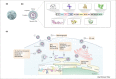
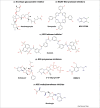
Despite the recent outbreak of Zika virus (ZIKV), there are still no approved treatments, and early-stage compounds are probably many years away from approval. A comprehensive A-Z review of the recent advances in ZIKV drug discovery efforts is presented, highlighting drug repositioning and computationally guided compounds, including discovered viral and host cell inhibitors. Promising ZIKV molecular targets are also described and discussed, as well as targets belonging to the host cell, as new opportunities for ZIKV drug discovery. All this knowledge is not only crucial to advancing the fight against the Zika virus and other flaviviruses but also helps us prepare for the next emerging virus outbreak to which we will have to respond.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures

References
-
- Hazin A.N. Computed tomographic findings in microcephaly associated with Zika virus. N. Engl. J. Med. 2016;374:2193–2195. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

