Alzheimer's disease master regulators analysis: search for potential molecular targets and drug repositioning candidates
- PMID: 29935546
- PMCID: PMC6015462
- DOI: 10.1186/s13195-018-0394-7
Alzheimer's disease master regulators analysis: search for potential molecular targets and drug repositioning candidates
Abstract
Background: Alzheimer's disease (AD) is a multifactorial and complex neuropathology that involves impairment of many intricate molecular mechanisms. Despite recent advances, AD pathophysiological characterization remains incomplete, which hampers the development of effective treatments. In fact, currently, there are no effective pharmacological treatments for AD. Integrative strategies such as transcription regulatory network and master regulator analyses exemplify promising new approaches to study complex diseases and may help in the identification of potential pharmacological targets.
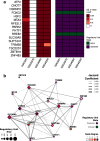
Methods: In this study, we used transcription regulatory network and master regulator analyses on transcriptomic data of human hippocampus to identify transcription factors (TFs) that can potentially act as master regulators in AD. All expression profiles were obtained from the Gene Expression Omnibus database using the GEOquery package. A normal hippocampus transcription factor-centered regulatory network was reconstructed using the ARACNe algorithm. Master regulator analysis and two-tail gene set enrichment analysis were employed to evaluate the inferred regulatory units in AD case-control studies. Finally, we used a connectivity map adaptation to prospect new potential therapeutic interventions by drug repurposing.
Results: We identified TFs with already reported involvement in AD, such as ATF2 and PARK2, as well as possible new targets for future investigations, such as CNOT7, CSRNP2, SLC30A9, and TSC22D1. Furthermore, Connectivity Map Analysis adaptation suggested the repositioning of six FDA-approved drugs that can potentially modulate master regulator candidate regulatory units (Cefuroxime, Cyproterone, Dydrogesterone, Metrizamide, Trimethadione, and Vorinostat).
Conclusions: Using a transcription factor-centered regulatory network reconstruction we were able to identify several potential molecular targets and six drug candidates for repositioning in AD. Our study provides further support for the use of bioinformatics tools as exploratory strategies in neurodegenerative diseases research, and also provides new perspectives on molecular targets and drug therapies for future investigation and validation in AD.
Keywords: Alzheimer’s disease; Drug repositioning; Hippocampus; Master regulators; Transcription factors; Transcriptional regulatory network reconstruction.
Conflict of interest statement
Ethics approval and consent to participate
Not applied.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Alzheimer’s Association. Alzheimer’s disease facts and figures. Alzheimers Dement. 2017;13:325–73.
-
- Prince MJ. World Alzheimer report 2015: the global impact of dementia: an analysis of prevalence, incidence, cost and trends. 2015.
-
- McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Jr, Kawas CH, Klunk WE, Koroshetz WJ, Manly JJ, Mayeux R, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

