Re-Expression of Bone Marrow Proteoglycan-2 by 5-Azacytidine is associated with STAT3 Inactivation and Sensitivity Response to Imatinib in Resistant CML Cells
- PMID: 29936783
- PMCID: PMC6103584
- DOI: 10.22034/APJCP.2018.19.6.1585
Re-Expression of Bone Marrow Proteoglycan-2 by 5-Azacytidine is associated with STAT3 Inactivation and Sensitivity Response to Imatinib in Resistant CML Cells
Abstract
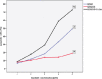
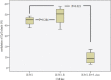
Background: Epigenetic silencing of tumor suppressor genes (TSG) is involved in development and progression of cancers. Re-expression of TSG is inversely proportionate with STAT3 signaling pathways. Demethylation of DNA by 5-Azacytidine (5-Aza) results in re-expression of silenced TSG. Forced expression of PRG2 by 5-Aza induced apoptosis in cancer cells. Imatinib is a tyrosine kinase inhibitor that potently inhibits BCR/ ABL tyrosine kinase resulting in hematological remission in CML patients. However, majority of CML patients treated with imatinib would develop resistance under prolonged therapy. Methods: CML cells resistant to imatinib were treated with 5-Aza and cytotoxicity of imatinib and apoptosis were determined by MTS and annexin-V, respectively. Gene expression analysis was detected by real time-PCR, STATs activity examined using Western blot and methylation status of PRG2 was determined by pyrosequencing analysis. Result: Expression of PRG2 was significantly higher in K562-R+5-Aza cells compared to K562 and K562-R (p=0.001). Methylation of PRG2 gene was significantly decreased in K562-R+5-Aza cells compared to other cells (p=0.021). STAT3 was inactivated in K562-R+5-Aza cells which showed higher sensitivity to imatinib. Conclusion: PRG2 gene is a TSG and its overexpression might induce sensitivity to imatinib. However, further studies are required to evaluate the negative regulations of PRG2 on STAT3 signaling.
Keywords: CML; PRG2; imatinib; 5-Azacytidine; STAT3.
Creative Commons Attribution License
Figures



References
-
- Al-jamal H, Jusoh S, Sidek M, et al. Restoration of PRG2 expression by 5-Azacytidine involves in sensitivity of PKC-412 (Midostaurin) resistant FLT3-ITD positiveacute Myeloid Leukaemia cells. J Hematol Thrombo Dis. 2015;3:2.
-
- Al-Jamal HA, Jusoh SA, Yong AC, et al. Silencing of suppressor of cytokine signaling-3 due to methylation results in phosphorylation of STAT3 in imatinib resistant BCR-ABL positive chronic myeloid leukemia cells. Asian Pac J Cancer Prev. 2014;15:4555–61. - PubMed
-
- Benekli M, Xia Z, Donohue KA, et al. Constitutive activity of signal transducer and activator of transcription 3 protein in acute myeloid leukemia blasts is associated with short disease-free survival. Blood. 2002;99:252–7. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

