LSD1 Ablation Stimulates Anti-tumor Immunity and Enables Checkpoint Blockade
- PMID: 29937226
- PMCID: PMC6063761
- DOI: 10.1016/j.cell.2018.05.052
LSD1 Ablation Stimulates Anti-tumor Immunity and Enables Checkpoint Blockade
Abstract
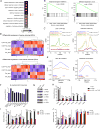
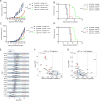
Chromatin regulators play a broad role in regulating gene expression and, when gone awry, can lead to cancer. Here, we demonstrate that ablation of the histone demethylase LSD1 in cancer cells increases repetitive element expression, including endogenous retroviral elements (ERVs), and decreases expression of RNA-induced silencing complex (RISC) components. Significantly, this leads to double-stranded RNA (dsRNA) stress and activation of type 1 interferon, which stimulates anti-tumor T cell immunity and restrains tumor growth. Furthermore, LSD1 depletion enhances tumor immunogenicity and T cell infiltration in poorly immunogenic tumors and elicits significant responses of checkpoint blockade-refractory mouse melanoma to anti-PD-1 therapy. Consistently, TCGA data analysis shows an inverse correlation between LSD1 expression and CD8+ T cell infiltration in various human cancers. Our study identifies LSD1 as a potent inhibitor of anti-tumor immunity and responsiveness to immunotherapy and suggests LSD1 inhibition combined with PD-(L)1 blockade as a novel cancer treatment strategy.
Keywords: LSD1; MHC-1; PD-1/PD-L1; RISC; T cell infiltration; anti-tumor immunity; dsRNA; endogenous retroviral element; immune checkpoint blockade; interferon.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
All other authors declare no competing interests.
Figures





References
-
- Brookes E, Shi Y. Diverse epigenetic mechanisms of human disease. Annu Rev Genet. 2014;48:237–268. - PubMed
-
- Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol. 2016;17:1142–1149. - PubMed
-
- Chen S, Lee LF, Fisher TS, Jessen B, Elliott M, Evering W, Logronio K, Tu GH, Tsaparikos K, Li X, et al. Combination of 4-1BB agonist and PD-1 antagonist promotes antitumor effector/memory CD8 T cells in a poorly immunogenic tumor model. Cancer Immunol Res. 2015;3:149–160. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

