Multiscale Analysis of Independent Alzheimer's Cohorts Finds Disruption of Molecular, Genetic, and Clinical Networks by Human Herpesvirus
- PMID: 29937276
- PMCID: PMC6551233
- DOI: 10.1016/j.neuron.2018.05.023
Multiscale Analysis of Independent Alzheimer's Cohorts Finds Disruption of Molecular, Genetic, and Clinical Networks by Human Herpesvirus
Abstract
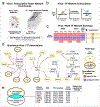
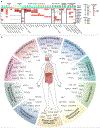
Investigators have long suspected that pathogenic microbes might contribute to the onset and progression of Alzheimer's disease (AD) although definitive evidence has not been presented. Whether such findings represent a causal contribution, or reflect opportunistic passengers of neurodegeneration, is also difficult to resolve. We constructed multiscale networks of the late-onset AD-associated virome, integrating genomic, transcriptomic, proteomic, and histopathological data across four brain regions from human post-mortem tissue. We observed increased human herpesvirus 6A (HHV-6A) and human herpesvirus 7 (HHV-7) from subjects with AD compared with controls. These results were replicated in two additional, independent and geographically dispersed cohorts. We observed regulatory relationships linking viral abundance and modulators of APP metabolism, including induction of APBB2, APPBP2, BIN1, BACE1, CLU, PICALM, and PSEN1 by HHV-6A. This study elucidates networks linking molecular, clinical, and neuropathological features with viral activity and is consistent with viral activity constituting a general feature of AD.
Keywords: Alzheimer's disease; HHV-6A; HHV-6B; HHV-7; Roseolovirus; human herpesvirus; integrative genomics; multiscale networks; network biology; systems biology.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declarations of Interests
The authors declare that they have no competing financial interests in relation to the work described.
Figures
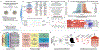

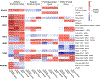

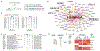
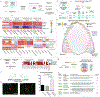
Comment in
-
Microbes and Alzheimer's Disease: New Findings Call for a Paradigm Change.Trends Neurosci. 2018 Sep;41(9):570-573. doi: 10.1016/j.tins.2018.07.001. Epub 2018 Jul 19. Trends Neurosci. 2018. PMID: 30033181
-
Commentary: Multiscale Analysis of Independent Alzheimer's Cohorts Finds Disruption of Molecular, Genetic, and Clinical Networks by Human Herpesvirus.Front Mol Neurosci. 2018 Sep 20;11:340. doi: 10.3389/fnmol.2018.00340. eCollection 2018. Front Mol Neurosci. 2018. PMID: 30294260 Free PMC article. No abstract available.
-
Are HHV-6A and HHV-7 Really More Abundant in Alzheimer's Disease?Neuron. 2019 Dec 18;104(6):1034-1035. doi: 10.1016/j.neuron.2019.11.009. Neuron. 2019. PMID: 31855626 No abstract available.
-
Clarifying the Potential Role of Microbes in Alzheimer's Disease.Neuron. 2019 Dec 18;104(6):1036-1037. doi: 10.1016/j.neuron.2019.11.008. Neuron. 2019. PMID: 31855627 Free PMC article. No abstract available.
-
Reanalysis of Alzheimer's brain sequencing data reveals absence of purported HHV6A and HHV7.J Bioinform Comput Biol. 2020 Feb;18(1):2050012. doi: 10.1142/S0219720020500122. J Bioinform Comput Biol. 2020. PMID: 32336252
References
-
- Agostini S,et al. (2016). Lack of evidence for a role of HHV-6 in the pathogenesis of Alzheimer’s disease. J Alzheimers Dis, 49, 229–35. - PubMed
-
- Albright AV,et al. (1998). The effect of human herpesvirus-6 (HHV-6) on cultured human neural cells: oligodendrocytes and microglia. J Neurovirol, 4, 486–94. - PubMed
-
- Altschul SF,et al. (1990). Basic local alignment search tool. J Mol Biol, 215, 403–10. - PubMed
-
- Ament S,et al. (2017). TReNA: Fit transcriptional regulatory networks using gene expression, priors, machine learning. R package version 0.99.10 ed.
Publication types
MeSH terms
Substances
Grants and funding
- RF1 AG058469/AG/NIA NIH HHS/United States
- U01 AG046152/AG/NIA NIH HHS/United States
- RF1 AG015819/AG/NIA NIH HHS/United States
- U54 EB020406/EB/NIBIB NIH HHS/United States
- P50 AG016574/AG/NIA NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- R01 AG032990/AG/NIA NIH HHS/United States
- R01 NS080820/NS/NINDS NIH HHS/United States
- U01 AG046139/AG/NIA NIH HHS/United States
- P01 AG017216/AG/NIA NIH HHS/United States
- R01 AG018023/AG/NIA NIH HHS/United States
- R56 AG058469/AG/NIA NIH HHS/United States
- U01 AG032984/AG/NIA NIH HHS/United States
- R01 AG030146/AG/NIA NIH HHS/United States
- U01 AG046170/AG/NIA NIH HHS/United States
- R01 AG017917/AG/NIA NIH HHS/United States
- R34 AG049649/AG/NIA NIH HHS/United States
- P01 AG003949/AG/NIA NIH HHS/United States
- U24 NS072026/NS/NINDS NIH HHS/United States
- P30 AG019610/AG/NIA NIH HHS/United States
- P50 AG025711/AG/NIA NIH HHS/United States
- U01 AG006786/AG/NIA NIH HHS/United States
- R01 AG036836/AG/NIA NIH HHS/United States
- R01 AG015819/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

