Deconstructing Adipogenesis Induced by β3-Adrenergic Receptor Activation with Single-Cell Expression Profiling
- PMID: 29937373
- PMCID: PMC6082711
- DOI: 10.1016/j.cmet.2018.05.025
Deconstructing Adipogenesis Induced by β3-Adrenergic Receptor Activation with Single-Cell Expression Profiling
Abstract
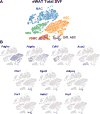
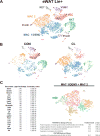
Recruitment of brown/beige adipocytes (BAs) in white adipose tissue (WAT) involves proliferation and differentiation of adipocyte stem cells (ASCs) in concert with close interactions with resident immune cells. To deconvolve stromal cell heterogeneity in a comprehensive and unbiased fashion, we performed single-cell RNA sequencing (scRNA-seq) of >33,000 stromal/vascular cells from epididymal WAT (eWAT) and inguinal WAT (iWAT) under control conditions and during β3-adrenergic receptor (ADRB3) activation. scRNA-seq identified distinct ASC subpopulations in eWAT and iWAT that appeared to be differentially poised to enter the adipogenic pathway. ADRB3 activation triggered the dramatic appearance of proliferating ASCs in eWAT, whose differentiation into BAs could be inferred from a single time point. scRNA-seq identified various immune cell types in eWAT, including a proliferating macrophage subpopulation that occupies adipogenic niches. These results demonstrate the power of scRNA-seq to deconstruct adipogenic niches and suggest novel functional interactions among resident stromal cell subpopulations.
Keywords: RNA-seq; adipocyte stem cell; adipogenesis; brown adipocyte; browning; differentiation trajectory; macrophage; recuitment; stromal cells.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures




References
-
- Barbatelli G, Murano I, Madsen L, Hao Q, Jimenez M, Kristiansen K, Giacobino JP, De Matteis R, Cinti S. The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte transdifferentiation. Am J Physiol Endocrinol Metab. 2010;298:E1244–1253. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

