The Drosophila Immune Deficiency Pathway Modulates Enteroendocrine Function and Host Metabolism
- PMID: 29937377
- PMCID: PMC6125180
- DOI: 10.1016/j.cmet.2018.05.026
The Drosophila Immune Deficiency Pathway Modulates Enteroendocrine Function and Host Metabolism
Abstract
Enteroendocrine cells (EEs) are interspersed between enterocytes and stem cells in the Drosophila intestinal epithelium. Like enterocytes, EEs express components of the immune deficiency (IMD) innate immune pathway, which activates transcription of genes encoding antimicrobial peptides. The discovery of large lipid droplets in intestines of IMD pathway mutants prompted us to investigate the role of the IMD pathway in the host metabolic response to its intestinal microbiota. Here we provide evidence that the short-chain fatty acid acetate is a microbial metabolic signal that activates signaling through the enteroendocrine IMD pathway in a PGRP-LC-dependent manner. This, in turn, increases transcription of the gene encoding the endocrine peptide Tachykinin (Tk), which is essential for timely larval development and optimal lipid metabolism and insulin signaling. Our findings suggest innate immune pathways not only provide the first line of defense against infection but also afford the intestinal microbiota control over host development and metabolism.
Keywords: Drosophila insulin-like peptide 3; PGRP-LC; enteroendocrine cell; enteroendocrine peptide; immune deficiency pathway; innate immunity; lipid droplet; metabolism; short-chain fatty acid; tachykinin.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures



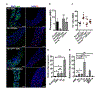
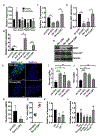


References
-
- Aggrawal K, and Silverman N (2007). Peptidoglycan recognition in Drosophila. Biochem Soc Trans 35, 1496–1500. - PubMed
-
- Bohni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, Beckingham K, and Hafen E (1999). Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1–4. Cell 97, 865–875. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

