Evolutionary History of the Glycoside Hydrolase 3 (GH3) Family Based on the Sequenced Genomes of 48 Plants and Identification of Jasmonic Acid-Related GH3 Proteins in Solanum tuberosum
- PMID: 29937487
- PMCID: PMC6073592
- DOI: 10.3390/ijms19071850
Evolutionary History of the Glycoside Hydrolase 3 (GH3) Family Based on the Sequenced Genomes of 48 Plants and Identification of Jasmonic Acid-Related GH3 Proteins in Solanum tuberosum
Abstract
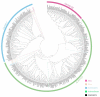
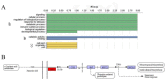
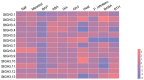
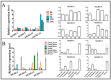
Glycoside Hydrolase 3 (GH3) is a phytohormone-responsive family of proteins found in many plant species. These proteins contribute to the biological activity of indolacetic acid (IAA), jasmonic acid (JA), and salicylic acid (SA). They also affect plant growth and developmental processes as well as some types of stress. In this study, GH3 genes were identified in 48 plant species, including algae, mosses, ferns, gymnosperms, and angiosperms. No GH3 representative protein was found in algae, but we identified 4 genes in mosses, 19 in ferns, 7 in gymnosperms, and several in angiosperms. The results showed that GH3 proteins are mainly present in seed plants. Phylogenetic analysis of all GH3 proteins showed three separate clades. Group I was related to JA adenylation, group II was related to IAA adenylation, and group III was separated from group II, but its function was not clear. The structure of the GH3 proteins indicated highly conserved sequences in the plant kingdom. The analysis of JA adenylation in relation to gene expression of GH3 in potato (Solanum tuberosum) showed that StGH3.12 greatly responded to methyl jasmonate (MeJA) treatment. The expression levels of StGH3.1, StGH3.11, and StGH3.12 were higher in the potato flowers, and StGH3.11 expression was also higher in the stolon. Our research revealed the evolution of the GH3 family, which is useful for studying the precise function of GH3 proteins related to JA adenylation in S. tuberosum when the plants are developing and under biotic stress.
Keywords: GH3 family; biotic; jasmonic acids; potato; sequencing plants; tissues.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Genome-wide identification, characterization analysis and expression profiling of auxin-responsive GH3 family genes in wheat (Triticum aestivum L.).Mol Biol Rep. 2020 May;47(5):3885-3907. doi: 10.1007/s11033-020-05477-5. Epub 2020 May 2. Mol Biol Rep. 2020. PMID: 32361896
-
Comprehensive Analysis of GH3 Gene Family in Potato and Functional Characterization of StGH3.3 under Drought Stress.Int J Mol Sci. 2023 Oct 12;24(20):15122. doi: 10.3390/ijms242015122. Int J Mol Sci. 2023. PMID: 37894803 Free PMC article.
-
Indole-3-acetic acid improves drought tolerance of white clover via activating auxin, abscisic acid and jasmonic acid related genes and inhibiting senescence genes.BMC Plant Biol. 2020 Apr 8;20(1):150. doi: 10.1186/s12870-020-02354-y. BMC Plant Biol. 2020. PMID: 32268884 Free PMC article.
-
Jasmonic Acid Signaling and Molecular Crosstalk with Other Phytohormones.Int J Mol Sci. 2021 Mar 13;22(6):2914. doi: 10.3390/ijms22062914. Int J Mol Sci. 2021. PMID: 33805647 Free PMC article. Review.
-
Crosstalk with Jasmonic Acid Integrates Multiple Responses in Plant Development.Int J Mol Sci. 2020 Jan 2;21(1):305. doi: 10.3390/ijms21010305. Int J Mol Sci. 2020. PMID: 31906415 Free PMC article. Review.
Cited by
-
Sedentary Plant-Parasitic Nematodes Alter Auxin Homeostasis via Multiple Strategies.Front Plant Sci. 2021 May 28;12:668548. doi: 10.3389/fpls.2021.668548. eCollection 2021. Front Plant Sci. 2021. PMID: 34122488 Free PMC article. Review.
-
Evolutionary significance of amino acid permease transporters in 17 plants from Chlorophyta to Angiospermae.BMC Genomics. 2020 Jun 5;21(1):391. doi: 10.1186/s12864-020-6729-3. BMC Genomics. 2020. PMID: 32503414 Free PMC article.
-
Functional characterization of Gh_A08G1120 (GH3.5) gene reveal their significant role in enhancing drought and salt stress tolerance in cotton.BMC Genet. 2019 Jul 23;20(1):62. doi: 10.1186/s12863-019-0756-6. BMC Genet. 2019. PMID: 31337336 Free PMC article.
-
Comparative Analysis of Hulless Barley Transcriptomes to Regulatory Effects of Phosphorous Deficiency.Life (Basel). 2024 Jul 19;14(7):904. doi: 10.3390/life14070904. Life (Basel). 2024. PMID: 39063656 Free PMC article.
-
Changes in Gene Expression in Leaves of Cacao Genotypes Resistant and Susceptible to Phytophthora palmivora Infection.Front Plant Sci. 2022 Feb 8;12:780805. doi: 10.3389/fpls.2021.780805. eCollection 2021. Front Plant Sci. 2022. PMID: 35211126 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

