Construction of a network describing asparagine metabolism in plants and its application to the identification of genes affecting asparagine metabolism in wheat under drought and nutritional stress
- PMID: 29938110
- PMCID: PMC5993343
- DOI: 10.1002/fes3.126
Construction of a network describing asparagine metabolism in plants and its application to the identification of genes affecting asparagine metabolism in wheat under drought and nutritional stress
Abstract
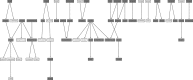
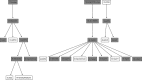
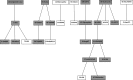
A detailed network describing asparagine metabolism in plants was constructed using published data from Arabidopsis (Arabidopsis thaliana) maize (Zea mays), wheat (Triticum aestivum), pea (Pisum sativum), soybean (Glycine max), lupin (Lupus albus), and other species, including animals. Asparagine synthesis and degradation is a major part of amino acid and nitrogen metabolism in plants. The complexity of its metabolism, including limiting and regulatory factors, was represented in a logical sequence in a pathway diagram built using yED graph editor software. The network was used with a Unique Network Identification Pipeline in the analysis of data from 18 publicly available transcriptomic data studies. This identified links between genes involved in asparagine metabolism in wheat roots under drought stress, wheat leaves under drought stress, and wheat leaves under conditions of sulfur and nitrogen deficiency. The network represents a powerful aid for interpreting the interactions not only between the genes in the pathway but also among enzymes, metabolites and smaller molecules. It provides a concise, clear understanding of the complexity of asparagine metabolism that could aid the interpretation of data relating to wider amino acid metabolism and other metabolic processes.
Keywords: asparagine metabolism; asparagine synthetase; glutamine synthetase; stress responses; systems approaches.
Figures


References
-
- Avila‐Ospina, L. , Marmagne, A. , Talbotec, J. , & Krupinska, K. (2015). The identification of new cytosolic glutamine synthetase and asparagine synthetase genes in barley (Hordeum vulgare L.), and their expression during leaf senescence. Journal of Experimental Botany, 66, 2013–2026. - PMC - PubMed
-
- Baena‐González, E. , Rolland, F. , Thevelein, J. M. , & Sheen, J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature, 448, 938–942. - PubMed
-
- Baker, J. M. , Hawkins, N. D. , Ward, J. L. , Lovegrove, A. , Napier, J. A. , Shewry, P. R. , & Beale, M. (2006). A metabolomic study of substantial equivalence of field‐grown genetically modified wheat. Plant Biotechnology Journal, 4, 381–392. - PubMed
-
- Baudet, J. , Huet, J.‐C. , Jolivet, E. , Lesaint, C. , Mossé, J. , & Pernollet, J.‐C. (1986). Changes in accumulation of seed nitrogen compounds in maize under conditions of sulphur deficiency. Physiologia Plantarum, 68, 608–614.
LinkOut - more resources
Full Text Sources
Other Literature Sources
