Application of the CRISPR/Cas9 System to Drug Resistance in Breast Cancer
- PMID: 29938175
- PMCID: PMC6010891
- DOI: 10.1002/advs.201700964
Application of the CRISPR/Cas9 System to Drug Resistance in Breast Cancer
Abstract
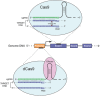
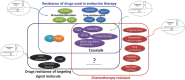
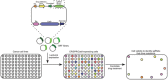
Clinical evidence indicates that drug resistance is a great obstacle in breast cancer therapy. It renders the disease uncontrollable and causes high mortality. Multiple mechanisms contribute to the development of drug resistance, but the underlying cause is usually a shift in the genetic composition of tumor cells. It is increasingly feasible to engineer the genome with the clustered regularly interspaced short palindromic repeats (CRISPR)/associated (Cas)9 technology recently developed, which might be advantageous in overcoming drug resistance. This article discusses how the CRISPR/Cas9 system might revert resistance gene mutations and identify potential resistance targets in drug-resistant breast cancer. In addition, the challenges that impede the clinical applicability of this technology and highlight the CRISPR/Cas9 systems are presented. The CRISPR/Cas9 system is poised to play an important role in preventing drug resistance in breast cancer therapy and will become an essential tool for personalized medicine.
Keywords: CRISPR/Cas9; breast cancer; drug resistance; drug therapy; reverting resistance.
Figures
Similar articles
-
Research into overcoming drug resistance in lung cancer treatment using CRISPR-Cas9 technology: a narrative review.Transl Lung Cancer Res. 2024 Aug 31;13(8):2067-2081. doi: 10.21037/tlcr-24-592. Epub 2024 Aug 28. Transl Lung Cancer Res. 2024. PMID: 39263032 Free PMC article. Review.
-
CRISPR-Cas9, A Promising Therapeutic Tool for Cancer Therapy: A Review.Protein Pept Lett. 2020;27(10):931-944. doi: 10.2174/0929866527666200407112432. Protein Pept Lett. 2020. PMID: 32264803
-
Applications of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) as a Genetic Scalpel for the Treatment of Cancer: A Translational Narrative Review.Cureus. 2023 Dec 6;15(12):e50031. doi: 10.7759/cureus.50031. eCollection 2023 Dec. Cureus. 2023. PMID: 38186450 Free PMC article. Review.
-
The application of CRISPR-Cas9 genome editing tool in cancer immunotherapy.Brief Funct Genomics. 2019 Mar 22;18(2):129-132. doi: 10.1093/bfgp/ely011. Brief Funct Genomics. 2019. PMID: 29579146 Review.
-
[Advances in application of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 system in stem cells research].Zhonghua Shao Shang Za Zhi. 2018 Apr 20;34(4):253-256. doi: 10.3760/cma.j.issn.1009-2587.2018.04.013. Zhonghua Shao Shang Za Zhi. 2018. PMID: 29690746 Review. Chinese.
Cited by
-
Systemic and Local Strategies for Primary Prevention of Breast Cancer.Cancers (Basel). 2024 Jan 5;16(2):248. doi: 10.3390/cancers16020248. Cancers (Basel). 2024. PMID: 38254741 Free PMC article. Review.
-
A Review on Nanocarrier Mediated Treatment and Management of Triple Negative Breast Cancer: A Saudi Arabian Scenario.Front Oncol. 2022 Jul 22;12:953865. doi: 10.3389/fonc.2022.953865. eCollection 2022. Front Oncol. 2022. PMID: 35941873 Free PMC article. Review.
-
Applications and challenges of CRISPR-Cas gene-editing to disease treatment in clinics.Precis Clin Med. 2021 Jul 10;4(3):179-191. doi: 10.1093/pcmedi/pbab014. eCollection 2021 Sep. Precis Clin Med. 2021. PMID: 34541453 Free PMC article. Review.
-
In Silico Analysis of Novel Bacterial Metabolites with Anticancer Activities.Metabolites. 2024 Mar 13;14(3):163. doi: 10.3390/metabo14030163. Metabolites. 2024. PMID: 38535323 Free PMC article.
-
A review of the literature on the use of CRISPR/Cas9 gene therapy to treat hepatocellular carcinoma.Oncol Res. 2024 Feb 6;32(3):439-461. doi: 10.32604/or.2023.044473. eCollection 2024. Oncol Res. 2024. PMID: 38361756 Free PMC article. Review.
References
-
- Turkoz F. P., Solak M., Petekkaya I., Keskin O., Kertmen N., Sarici F., Arik Z., Babacan T., Ozisik Y., Altundag K., Breast 2013, 22, 344. - PubMed
-
- Curigliano G., Cancer Treat. Rev. 2012, 38, 303. - PubMed
-
- Vogel C. L., Johnston M. A., Capers C., Braccia D., Clin. Breast Cancer 2014, 14, 1. - PubMed
-
- Giuliano M., Hu H., Wang Y. C., Fu X., Nardone A., Herrera S., Mao S., Contreras A., Gutierrez C., Wang T., Hilsenbeck S. G., De Angelis C., Wang N. J., Heiser L. M., Gray J. W., Lopez‐Tarruella S., Pavlick A. C., Trivedi M. V., Chamness G. C., Chang J. C., Osborne C. K., Rimawi M. F., Schiff R., Clin. Cancer Res. 2015, 21, 3995. - PMC - PubMed
-
- Zilli M., Grassadonia A., Tinari N., Di Giacobbe A., Gildetti S., Giampietro J., Natoli C., Iacobelli S., Biochim. Biophys. Acta 2009, 1795, 62. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
