Runx1 Role in Epithelial and Cancer Cell Proliferation Implicates Lipid Metabolism and Scd1 and Soat1 Activity
- PMID: 29938858
- PMCID: PMC6202256
- DOI: 10.1002/stem.2868
Runx1 Role in Epithelial and Cancer Cell Proliferation Implicates Lipid Metabolism and Scd1 and Soat1 Activity
Abstract
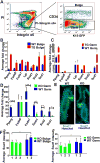
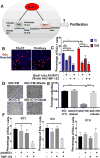
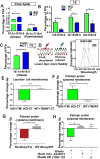
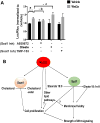
The role of lipid metabolism in epithelial stem cell (SC) function and carcinogenesis is poorly understood. The transcription factor Runx1 is known to regulate proliferation in mouse epithelial hair follicle (HF) SCs in vivo and in several mouse and human epithelial cancers. We found a novel subset of in vivo Runx1 HFSC target genes related to lipid metabolism and demonstrated changes in distinct classes of lipids driven by Runx1. Inhibition of lipid-enzymes Scd1 and Soat1 activity synergistically reduces proliferation of mouse skin epithelial cells and of human skin and oral squamous cell carcinoma cultured lines. Varying Runx1 levels induces changes in skin monounsaturated fatty acids (e.g., oleate, a product of Scd1) as shown by our lipidome analysis. Furthermore, varying Runx1 levels, the inhibition of Scd1, or the addition of Scd1-product oleate, individually affects the plasma membrane organization (or fluidity) in mouse keratinocytes. These factors also affect the strength of signal transduction through the membranes for Wnt, a pathway that promotes epithelial (cancer) cell proliferation and HFSC activation. Our working model is that HFSC factor Runx1 modulates the fatty acid production, which affects membrane organization, facilitating signal transduction for rapid proliferation of normal and cancer epithelial cells. Stem Cells 2018;36:1603-1616.
Keywords: Adult stem cells; Cancer; Cellular proliferation; Epidermis; Signal transduction; fluorescence-activated cell sorter.
© AlphaMed Press 2018.
Conflict of interest statement
Authors declare no conflict of interests.
Figures


Similar articles
-
Runx1 directly promotes proliferation of hair follicle stem cells and epithelial tumor formation in mouse skin.Mol Cell Biol. 2010 May;30(10):2518-36. doi: 10.1128/MCB.01308-09. Epub 2010 Mar 22. Mol Cell Biol. 2010. PMID: 20308320 Free PMC article.
-
Defining a tissue stem cell-driven Runx1/Stat3 signalling axis in epithelial cancer.EMBO J. 2012 Nov 5;31(21):4124-39. doi: 10.1038/emboj.2012.270. Epub 2012 Oct 2. EMBO J. 2012. PMID: 23034403 Free PMC article.
-
Runx1 and p21 synergistically limit the extent of hair follicle stem cell quiescence in vivo.Proc Natl Acad Sci U S A. 2013 Mar 19;110(12):4634-9. doi: 10.1073/pnas.1213015110. Epub 2013 Mar 4. Proc Natl Acad Sci U S A. 2013. PMID: 23487742 Free PMC article.
-
New insights into the role of Runx1 in epithelial stem cell biology and pathology.J Cell Biochem. 2013 May;114(5):985-93. doi: 10.1002/jcb.24453. J Cell Biochem. 2013. PMID: 23150456 Free PMC article. Review.
-
Stearoyl-CoA desaturase--a new player in skeletal muscle metabolism regulation.J Physiol Pharmacol. 2006 Nov;57 Suppl 10:31-42. J Physiol Pharmacol. 2006. PMID: 17242489 Review.
Cited by
-
RUNX1 Dosage in Development and Cancer.Mol Cells. 2020 Feb 29;43(2):126-138. doi: 10.14348/molcells.2019.0301. Mol Cells. 2020. PMID: 31991535 Free PMC article. Review.
-
Complex Interplay between the RUNX Transcription Factors and Wnt/β-Catenin Pathway in Cancer: A Tango in the Night.Mol Cells. 2020 Feb 29;43(2):188-197. doi: 10.14348/molcells.2019.0310. Mol Cells. 2020. PMID: 32041394 Free PMC article. Review.
-
Stem cell-intrinsic mechanisms regulating adult hair follicle homeostasis.Exp Dermatol. 2021 Apr;30(4):430-447. doi: 10.1111/exd.14251. Epub 2020 Dec 20. Exp Dermatol. 2021. PMID: 33278851 Free PMC article. Review.
-
RUNX2 interacts with SCD1 and activates Wnt/β-catenin signaling pathway to promote the progression of clear cell renal cell carcinoma.Cancer Med. 2023 Mar;12(5):5764-5780. doi: 10.1002/cam4.5326. Epub 2022 Oct 6. Cancer Med. 2023. PMID: 36200301 Free PMC article.
-
Lipid metabolism in cancer progression and therapeutic strategies.MedComm (2020). 2020 Dec 24;2(1):27-59. doi: 10.1002/mco2.27. eCollection 2021 Mar. MedComm (2020). 2020. PMID: 34766135 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

