Expansion microscopy: enabling single cell analysis in intact biological systems
- PMID: 29938896
- PMCID: PMC6309752
- DOI: 10.1111/febs.14597
Expansion microscopy: enabling single cell analysis in intact biological systems
Abstract
There is a need for single cell analysis methods that enable the identification and localization of different kinds of biomolecules throughout cells and intact tissues, thereby allowing characterization and classification of individual cells and their relationships to each other within intact systems. Expansion microscopy (ExM) is a technology that physically magnifies tissues in an isotropic way, thereby achieving super-resolution microscopy on diffraction-limited microscopes, enabling rapid image acquisition and large field of view. As a result, ExM is well-positioned to integrate molecular content and cellular morphology, with the spatial precision sufficient to resolve individual biological building blocks, and the scale and accessibility required to deploy over extended 3-D objects like tissues and organs.
Keywords: FISH; expansion microscopy; genomics; morphology; multiplexing; single cell analysis; super-resolution microscopy.
© 2018 Federation of European Biochemical Societies.
Conflict of interest statement
Competing financial interests
ESB is a co-inventor on multiple patents relating to ExM, and is also a co-founder of a company (
Figures

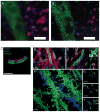
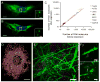
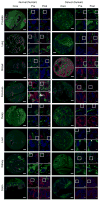
References
-
- Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell P, Carninci P, Clatworthy M, Clevers H, Deplancke B, Dunham I, Eberwine J, Eils R, Enard W, Farmer A, Fugger L, Göttgens B, Hacohen N, Haniffa M, Hemberg M, Kim S, Klenerman P, Kriegstein A, Lein E, Linnarsson S, Lundberg E, Lundeberg J, Majumder P, Marioni JC, Merad M, Mhlanga M, Nawijn M, Netea M, Nolan G, Pe’er D, Phillipakis A, Ponting CP, Quake S, Reik W, Rozenblatt-Rosen O, Sanes J, Satija R, Schumacher TN, Shalek A, Shapiro E, Sharma P, Shin JW, Stegle O, Stratton M, Stubbington MJT, Theis FJ, Uhlen M, van Oudenaarden A, Wagner A, Watt F, Weissman J, Wold B, Xavier R, Yosef N Human Cell Atlas Meeting Participants. The Human Cell Atlas. Elife. 2017:6. - PMC - PubMed
-
- Zeng H, Sanes JR. Neuronal cell-type classification: challenges, opportunities and the path forward. Nat Rev Neurosci 2017 - PubMed
-
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. - PMC - PubMed
-
- Eberwine J, Sul J-Y, Bartfai T, Kim J. The promise of single-cell sequencing. Nat Methods. 2013;11:25–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

