Oxytocin-like signaling in ants influences metabolic gene expression and locomotor activity
- PMID: 29939785
- PMCID: PMC6174076
- DOI: 10.1096/fj.201800443
Oxytocin-like signaling in ants influences metabolic gene expression and locomotor activity
Abstract
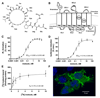
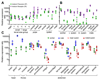
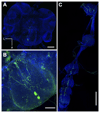
Ants are emerging model systems to study cellular signaling because distinct castes possess different physiologic phenotypes within the same colony. Here we studied the functionality of inotocin signaling, an insect ortholog of mammalian oxytocin (OT), which was recently discovered in ants. In Lasius ants, we determined that specialization within the colony, seasonal factors, and physiologic conditions down-regulated the expression of the OT-like signaling system. Given this natural variation, we interrogated its function using RNAi knockdowns. Next-generation RNA sequencing of OT-like precursor knock-down ants highlighted its role in the regulation of genes involved in metabolism. Knock-down ants exhibited higher walking activity and increased self-grooming in the brood chamber. We propose that OT-like signaling in ants is important for regulating metabolic processes and locomotion.-Liutkevičiūtė, Z., Gil-Mansilla, E., Eder, T., Casillas-Pérez, B., Di Giglio, M. G., Muratspahić, E., Grebien, F., Rattei, T., Muttenthaler, M., Cremer, S., Gruber, C. W. Oxytocin-like signaling in ants influences metabolic gene expression and locomotor activity.
Keywords: G protein–coupled receptor; inotocin; insect; vasopressin; vasotocin.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
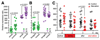



References
-
- Gäde G. Regulation of intermediary metabolism and water balance of insects by neuropeptides. Annu Rev Entomol. 2004;49:93–113. - PubMed
-
- Donaldson ZR, Young LJ. Oxytocin, vasopressin, and the neurogenetics of sociality. Science. 2008;322:900–904. - PubMed
-
- Gimpl G, Fahrenholz F. The oxytocin receptor system: structure, function, and regulation. Physiol Rev. 2001;81:629–683. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

