Exploring the single-cell RNA-seq analysis landscape with the scRNA-tools database
- PMID: 29939984
- PMCID: PMC6034903
- DOI: 10.1371/journal.pcbi.1006245
Exploring the single-cell RNA-seq analysis landscape with the scRNA-tools database
Abstract
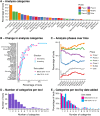
As single-cell RNA-sequencing (scRNA-seq) datasets have become more widespread the number of tools designed to analyse these data has dramatically increased. Navigating the vast sea of tools now available is becoming increasingly challenging for researchers. In order to better facilitate selection of appropriate analysis tools we have created the scRNA-tools database (www.scRNA-tools.org) to catalogue and curate analysis tools as they become available. Our database collects a range of information on each scRNA-seq analysis tool and categorises them according to the analysis tasks they perform. Exploration of this database gives insights into the areas of rapid development of analysis methods for scRNA-seq data. We see that many tools perform tasks specific to scRNA-seq analysis, particularly clustering and ordering of cells. We also find that the scRNA-seq community embraces an open-source and open-science approach, with most tools available under open-source licenses and preprints being extensively used as a means to describe methods. The scRNA-tools database provides a valuable resource for researchers embarking on scRNA-seq analysis and records the growth of the field over time.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures


References
-
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6: 377–382. doi: 10.1038/nmeth.1315 - DOI - PubMed
-
- Svensson V, Vento-Tormo R, Teichmann SA. Exponential scaling of single-cell RNA-seq in the past decade. Nat Protoc. 2018;13: 599 doi: 10.1038/nprot.2017.149 - DOI - PubMed
-
- Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16: 133–145. doi: 10.1038/nrg3833 - DOI - PubMed
-
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12: 115–121. doi: 10.1038/nmeth.3252 - DOI - PMC - PubMed
-
- Chamberlain S, Boettiger C, Hart T, Ram K. rcrossref: Client for Various ‘CrossRef’ ‘APIs’. 2017. https://CRAN.R-project.org/package=rcrossref
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

