Non-linear hierarchy of the quorum sensing signalling pathway in bloodstream form African trypanosomes
- PMID: 29940034
- PMCID: PMC6034907
- DOI: 10.1371/journal.ppat.1007145
Non-linear hierarchy of the quorum sensing signalling pathway in bloodstream form African trypanosomes
Abstract
Trypanosoma brucei, the agents of African trypanosomiasis, undergo density-dependent differentiation in the mammalian bloodstream to prepare for transmission by tsetse flies. This involves the generation of cell-cycle arrested, quiescent, stumpy forms from proliferative slender forms. The signalling pathway responsible for the quorum sensing response has been catalogued using a genome-wide selective screen, providing a compendium of signalling protein kinases phosphatases, RNA binding proteins and hypothetical proteins. However, the ordering of these components is unknown. To piece together these components to provide a description of how stumpy formation arises we have used an extragenic suppression approach. This exploited a combinatorial gene knockout and overexpression strategy to assess whether the loss of developmental competence in null mutants of pathway components could be compensated by ectopic expression of other components. We have created null mutants for three genes in the stumpy induction factor signalling pathway (RBP7, YAK, MEKK1) and evaluated complementation by expression of RBP7, NEK17, PP1-6, or inducible gene silencing of the proposed differentiation inhibitor TbTOR4. This indicated that the signalling pathway is non-linear. Phosphoproteomic analysis focused on one pathway component, a putative MEKK, identified molecules with altered expression and phosphorylation profiles in MEKK1 null mutants, including another component in the pathway, NEK17. Our data provide a first molecular dissection of multiple components in a signal transduction cascade in trypanosomes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

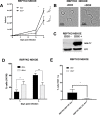
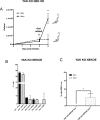
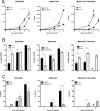

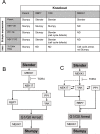
References
-
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annual review of cell and developmental biology. 2005;21:319–46. Epub 2005/10/11. doi: 10.1146/annurev.cellbio.21.012704.131001 . - DOI - PubMed
-
- Druker BJ, Mamon HJ, Roberts TM. Oncogenes, growth factors, and signal transduction. N Engl J Med. 1989;321(20):1383–91. Epub 1989/11/16. doi: 10.1056/NEJM198911163212007 . - DOI - PubMed
-
- Marsh L, Neiman AM, Herskowitz I. Signal transduction during pheromone response in yeast. Annu Rev Cell Biol. 1991;7:699–728. Epub 1991/01/01. doi: 10.1146/annurev.cb.07.110191.003411 . - DOI - PubMed
-
- Parent CA, Devreotes PN. Molecular genetics of signal transduction in Dictyostelium. Annual Review of Biochemistry. 1996;65:411–40. doi: 10.1146/annurev.bi.65.070196.002211 - DOI - PubMed
-
- O'Farrell PH. Quiescence: early evolutionary origins and universality do not imply uniformity. Philos Trans R Soc Lond B Biol Sci. 2011;366(1584):3498–507. doi: 10.1098/rstb.2011.0079 ; PubMed Central PMCID: PMCPMC3203459. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

