Inhibiting autophagy reduces retinal degeneration caused by protein misfolding
- PMID: 29940785
- PMCID: PMC6103695
- DOI: 10.1080/15548627.2018.1463121
Inhibiting autophagy reduces retinal degeneration caused by protein misfolding
Abstract
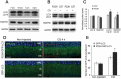
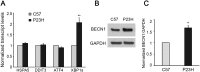
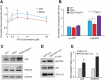
Mutations in the genes necessary for the structure and function of vertebrate photoreceptor cells are associated with multiple forms of inherited retinal degeneration. Mutations in the gene encoding RHO (rhodopsin) are a common cause of autosomal dominant retinitis pigmentosa (adRP), with the Pro23His variant of RHO resulting in a misfolded protein that activates endoplasmic reticulum stress and the unfolded protein response. Stimulating macroautophagy/autophagy has been proposed as a strategy for clearing misfolded RHO and reducing photoreceptor death. We found that retinas from mice heterozygous for the gene encoding the RHOP23H variant (hereafter called P23H) exhibited elevated levels of autophagy flux, and that pharmacological stimulation of autophagy accelerated retinal degeneration. In contrast, reducing autophagy flux pharmacologically or by rod-specific deletion of the autophagy-activating gene Atg5, improved photoreceptor structure and function. Furthermore, proteasome levels and activity were reduced in the P23H retina, and increased when Atg5 was deleted. Our findings suggest that autophagy contributes to photoreceptor cell death in P23H mice, and that decreasing autophagy shifts the degradation of misfolded RHO protein to the proteasome and is protective. These observations suggest that modulating the flux of misfolded proteins from autophagy to the proteasome may represent an important therapeutic strategy for reducing proteotoxicity in adRP and other diseases caused by protein folding defects.
Keywords: Autophagy; Rhodopsin; proteasome; protein misfolding; retinal degeneration.
Figures






Similar articles
-
Shifting the balance of autophagy and proteasome activation reduces proteotoxic cell death: a novel therapeutic approach for restoring photoreceptor homeostasis.Cell Death Dis. 2019 Jul 18;10(8):547. doi: 10.1038/s41419-019-1780-1. Cell Death Dis. 2019. PMID: 31320609 Free PMC article.
-
Pharmacological Inhibition of the VCP/Proteasome Axis Rescues Photoreceptor Degeneration in RHOP23H Rat Retinal Explants.Biomolecules. 2021 Oct 16;11(10):1528. doi: 10.3390/biom11101528. Biomolecules. 2021. PMID: 34680161 Free PMC article.
-
Calpain Activation Is the Major Cause of Cell Death in Photoreceptors Expressing a Rhodopsin Misfolding Mutation.Mol Neurobiol. 2020 Feb;57(2):589-599. doi: 10.1007/s12035-019-01723-5. Epub 2019 Aug 10. Mol Neurobiol. 2020. PMID: 31401765
-
Molecular genetics of retinal degeneration: A Drosophila perspective.Fly (Austin). 2011 Oct-Dec;5(4):356-68. doi: 10.4161/fly.5.4.17809. Epub 2011 Sep 7. Fly (Austin). 2011. PMID: 21897116 Free PMC article. Review.
-
Aberrant protein trafficking in retinal degenerations: The initial phase of retinal remodeling.Exp Eye Res. 2016 Sep;150:71-80. doi: 10.1016/j.exer.2015.11.007. Epub 2015 Nov 26. Exp Eye Res. 2016. PMID: 26632497 Free PMC article. Review.
Cited by
-
Beyond Genetics: The Role of Metabolism in Photoreceptor Survival, Development and Repair.Front Cell Dev Biol. 2022 May 18;10:887764. doi: 10.3389/fcell.2022.887764. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35663397 Free PMC article. Review.
-
HDAC inhibition ameliorates cone survival in retinitis pigmentosa mice.Cell Death Differ. 2021 Apr;28(4):1317-1332. doi: 10.1038/s41418-020-00653-3. Epub 2020 Nov 6. Cell Death Differ. 2021. PMID: 33159184 Free PMC article.
-
Haploinsufficiency of GCP4 induces autophagy and leads to photoreceptor degeneration due to defective spindle assembly in retina.Cell Death Differ. 2020 Feb;27(2):556-572. doi: 10.1038/s41418-019-0371-0. Epub 2019 Jun 17. Cell Death Differ. 2020. PMID: 31209365 Free PMC article.
-
Pharmacological clearance of misfolded rhodopsin for the treatment of RHO-associated retinitis pigmentosa.FASEB J. 2020 Aug;34(8):10146-10167. doi: 10.1096/fj.202000282R. Epub 2020 Jun 14. FASEB J. 2020. PMID: 32536017 Free PMC article.
-
Clinical trial design for neuroprotection in RHO autosomal dominant retinitis pigmentosa; outcome measure considerations.Ophthalmic Genet. 2021 Apr;42(2):170-177. doi: 10.1080/13816810.2020.1867752. Epub 2021 Jan 6. Ophthalmic Genet. 2021. PMID: 33406961 Free PMC article.
References
-
- RetNet – retinal information network. Accepted on 18 September 2017 Available from: https://sph.uth.edu/retnet/
-
- Saliba RS, Munro PM, Luthert PJ, et al. The cellular fate of mutant rhodopsin: quality control, degradation and aggresome formation. J Cell Sci. 2002;115:2907–2918. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
