IRE1B degrades RNAs encoding proteins that interfere with the induction of autophagy by ER stress in Arabidopsis thaliana
- PMID: 29940799
- PMCID: PMC6135571
- DOI: 10.1080/15548627.2018.1462426
IRE1B degrades RNAs encoding proteins that interfere with the induction of autophagy by ER stress in Arabidopsis thaliana
Abstract
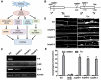
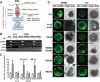
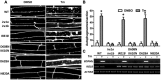
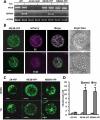
Macroautophagy/autophagy is a conserved process in eukaryotes that contributes to cell survival in response to stress. Previously, we found that endoplasmic reticulum (ER) stress induces autophagy in plants via a pathway dependent upon AT5G24360/IRE1B (INOSITOL REQUIRING 1-1), an ER membrane-anchored factor involved in the splicing of AT1G42990/BZIP60 (basic leucine zipper protein 60) mRNA. IRE1B is a dual protein kinase and ribonuclease, and here we determined the involvement of the protein kinase catalytic domain, nucleotide binding and RNase domains of IRE1B in activating autophagy. We found that the nucleotide binding and RNase activity of IRE1B, but not its protein kinase activity or splicing target BZIP60, are required for ER stress-mediated autophagy. Upon ER stress, the RNase activity of IRE1B engages in regulated IRE1-dependent decay of messenger RNA (RIDD), in which mRNAs of secreted proteins are degraded by IRE1 upon ER stress. Twelve genes most highly targeted by RIDD were tested for their role in inhibiting ER stress-induced autophagy, and 3 of their encoded proteins, AT1G66270/BGLU21 (β-glucosidase 21), AT2G16005/ROSY1/ML (MD2-related lipid recognition protein) and AT5G01870/PR-14 (pathogenesis-related protein 14), were found to inhibit autophagy upon overexpression. From these findings, IRE1B is posited to be a 'licensing factor' linking ER stress to autophagy by degrading the RNA transcripts of factors that interfere with the induction of autophagy.
Abbreviations: ACT2: actin 2; ATG: autophagy-related; BGLU21: β-glucosidase 21; BIP3: binding protein 3; BZIP: basic leucine zipper; DAPI: 4', 6-diamidino-2-phenylindole; DTT: dithiothreitol; ER: endoplasmic reticulum; ERN1: endoplasmic reticulum to nucleus signaling 1; IRE1: inositol requiring 1; GFP: green fluorescent protein; MAP3K5/ASK1: mitogen-activated protein kinase kinase kinase 5; MAPK8/JNK1: mitogen-activated protein kinase 8/c-Jun N-terminal kinase 1; MDC: monodansylcadaverine; PR-14: pathogenesis-related protein 14; RIDD: Regulated IRE1-Dependent Decay of Messenger RNA; ROSY1/ML: interactor of synaptotagmin1/MD2-related lipid recognition protein; Tm: tunicamycin; UPR: unfolded protein response; WT: wild-type.
Keywords: Arabidopsis; ER stress; IRE1; RIDD; autophagosome; autophagy; mRNA degradation.
Figures
References
-
- Boyer J. Plant productivity and environment. Science. 1982;218:443–448. - PubMed
-
- Yang X, Bassham DC. New insight into the mechanism and function of autophagy in plant cells. Int Rev Cell Mol Biol. 2015;320:1–40. Epub 2015/ 11/29. - PubMed
-
- Michaeli S, Galili G, Genschik P, et al. Autophagy in plants–what’s new on the menu? Trends Plant Sci. 2016;21:134–144. Epub 2015/ 11/26. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
