Proteome-wide analysis of human motif-domain interactions mapped on influenza a virus
- PMID: 29940841
- PMCID: PMC6019528
- DOI: 10.1186/s12859-018-2237-8
Proteome-wide analysis of human motif-domain interactions mapped on influenza a virus
Abstract
Background: The influenza A virus (IAV) is a constant threat for humans worldwide. The understanding of motif-domain protein participation is essential to combat the pathogen.
Results: In this study, a data mining approach was employed to extract influenza-human Protein-Protein interactions (PPI) from VirusMentha,Virus MINT, IntAct, and Pfam databases, to mine motif-domain interactions (MDIs) stored as Regular Expressions (RegExp) in 3DID database. A total of 107 RegExp related to human MDIs were searched on 51,242 protein fragments from H1N1, H1N2, H2N2, H3N2 and H5N1 strains obtained from Virus Variation database. A total 46 MDIs were frequently mapped on the IAV proteins and shared between the different strains. IAV kept host-like MDIs that were associated with the virus survival, which could be related to essential biological process such as microtubule-based processes, regulation of cell cycle check point, regulation of replication and transcription of DNA, etc. in human cells. The amino acid motifs were searched for matches in the immune epitope database and it was found that some motifs are part of experimentally determined epitopes on IAV, implying that such interactions exist.
Conclusion: The directed data-mining method employed could be used to identify functional motifs in other viruses for envisioning new therapies.
Keywords: Data-mining; Influenza; Motif-domain interactions; Protein-protein interactions.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
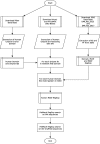

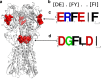
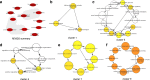
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

