Conservation status and historical relatedness of Italian cattle breeds
- PMID: 29940848
- PMCID: PMC6019226
- DOI: 10.1186/s12711-018-0406-x
Conservation status and historical relatedness of Italian cattle breeds
Abstract
Background: In the last 50 years, the diversity of cattle breeds has experienced a severe contraction. However, in spite of the growing diffusion of cosmopolite specialized breeds, several local cattle breeds are still farmed in Italy. Genetic characterization of breeds represents an essential step to guide decisions in the management of farm animal genetic resources. The aim of this work was to provide a high-resolution representation of the genome-wide diversity and population structure of Italian local cattle breeds using a medium-density single nucleotide polymorphism (SNP) array.
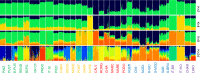
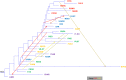
Results: After quality control filtering, the dataset included 31,013 SNPs for 800 samples from 32 breeds. Our results on the genetic diversity of these breeds agree largely with their recorded history. We observed a low level of genetic diversity, which together with the small size of the effective populations, confirmed that several breeds are threatened with extinction. According to the analysis of runs of homozygosity, evidence of recent inbreeding was strong in some local breeds, such as Garfagnina, Mucca Pisana and Pontremolese. Patterns of genetic differentiation, shared ancestry, admixture events, and the phylogenetic tree, all suggest the presence of gene flow, in particular among breeds that originate from the same geographical area, such as the Sicilian breeds. In spite of the complex admixture events that most Italian cattle breeds have experienced, they have preserved distinctive characteristics and can be clearly discriminated, which is probably due to differences in genetic origin, environment, genetic isolation and inbreeding.
Conclusions: This study is the first exhaustive genome-wide analysis of the diversity of Italian cattle breeds. The results are of significant importance because they will help design and implement conservation strategies. Indeed, efforts to maintain genetic diversity in these breeds are needed. Improvement of systems to record and monitor inbreeding in these breeds may contribute to their in situ conservation and, in view of this, the availability of genomic data is a fundamental resource.
Figures




References
-
- Felius M, Beerling ML, Buchanan DS, Theunissen B, Koolmees PA, Lenstra JA. On the history of cattle genetic resources. Diversity. 2014;6:705–750. doi: 10.3390/d6040705. - DOI
-
- Kukučková V, Moravčíková N, Ferenčaković M, Simčič M, Mészáros G, Sölkner J, et al. Genomic characterization of Pinzgau cattle: genetic conservation and breeding perspectives. Conserv Genet. 2017;18:839–910.
-
- Hiemstra SJ, de Haas Y, Mäki-Tanila A, Gandini G. Local cattle breeds in Europe—development of policies and strategies for self-sustaining breeds. Wageningen: Wageningen Academic Publishers; 2010.

